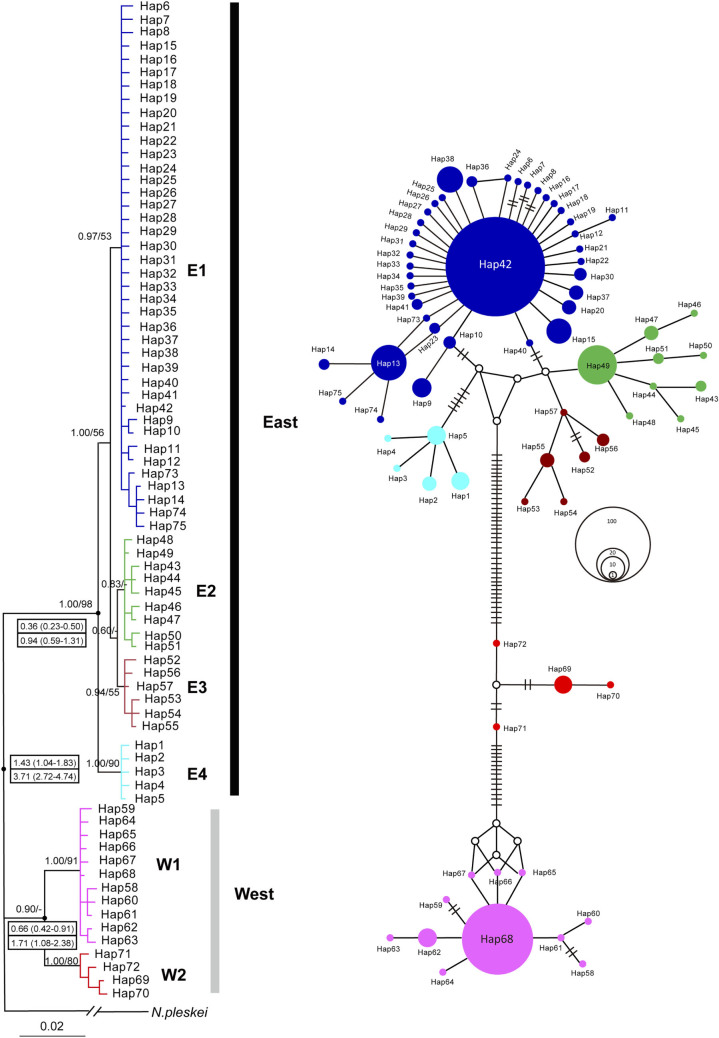
Figure 2. Bayesian phylogenetic tree (left) and the median-joining network (right) of mitochondrial Cytb haplotypes for N. parkeri.
Bayesian posterior probabilities/maximum likelihood bootstrap support values are above the branches. Mean time to the most recent common ancestor (TMRCA) with 95% highest posterior density (95% HPD) in parenthesis for the key nodes are given in the boxes (Mya): the above values are derived from the higher mutation rate (1.8 × 10-8/site/year) and the low from the mutation rate of 6.9 × 10-9/site/year. Colours in the network represent different Cytb lineages. In the network, sizes of cycles indicate the haplotype frequencies. Network branches linking the cycles indicate one mutation step; two or more mutations are represented by slashes crossed with the network branches. The small open circles are hypothetical missing intermediates.