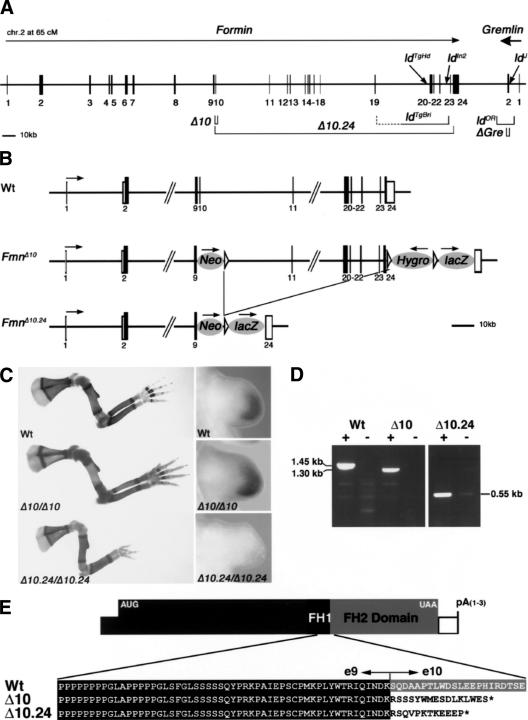
Figure 2.
Not disruption of the Formin FH2 domain, but deletion of the corresponding genomic region causes the ld limb phenotype. (A) Schematic representation of the ld complementation group consisting of Formin and Gremlin loci. The Formin gene is encoded by at least 24 exons (transcriptional direction indicated by arrow; Wang et al. 1997), whereas the Gre gene is transcribed in reverse orientation (bold arrow) and contains only two exons. The intergenic region separating the two genes is ∼38 kb. The Formin FH2 domain is encoded by exons 10–24 and is present in all Formin protein isoforms (Wang et al. 1997). The following genetically engineered mutations are indicated: (Δ10) FmnΔ10 allele; (Δ10.24) FmnΔ10.24 allele; (ΔGre) GreΔORF null allele (Michos et al. 2004). The spontaneous ld alleles are indicated: (ldTgBri) transgene induced deletion of genomic region between exons 19 and 23 (Vogt et al. 1992); (ldTgHd) transgene insertional mutagenesis (Woychik et al. 1985); (ldIn2) 40-Mb inversion involving Formin and Agouti loci (Woychik et al. 1990); (ldOR) deletion of the Gre ORF; (ldJ) point mutation disrupting Gre pre-mRNA splicing. (B) Schematic representation of the genetically engineered FmnΔ10 and FmnΔ10.24 alleles. (Neo) PGK-NeoR gene used to select ES-cell clones (first round of gene targeting); (Hygro) PGK-HygroR gene used to select ES-cell clones (second round of gene targeting); (lacZ) IRES-LacZ gene used to tag Fmn transcripts. Arrows indicate direction of transcription. Formin exons are numbered as in A. (C, left panels) Limb skeletal phenotypes of wild-type and homozygous mice. Genotypes are indicated in the panels. (Right panels) Gremlin expression in limb buds of wild-type and homozygous embryos (E10.75). For nomenclature see the legend for A. (D) RT–PCR of Formin transcripts isolated from wild-type (Wt), FmnΔ10 (Δ10) and FmnΔ10.24 (Δ10.24) homozygous embryos. Wild-type and FmnΔ10 mRNAs extending downstream from exon 9 were detected using primers in exons 9 and 23, FmnΔ10.24 mRNAs extending downstream from exon 9 were detected using primers in exon 9 and the IRES-LacZ tag (see Materials and Methods). (+) Reverse transcriptase included; (-) reverse transcriptase omitted (control). Note that the difference in size between wild-type (Wt) and FmnΔ10 transcripts is 150 bases, as expected. (E) Amino acid sequence deduced from the sequences of the Formin transcripts arising from wild-type, FmnΔ10 and FmnΔ10.24 alleles.