Figure 2.
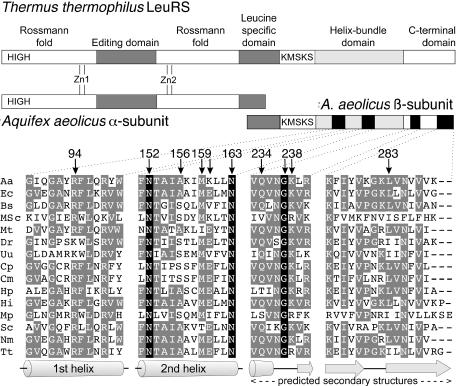
Schematic diagram of the domain structure of T.thermophilus and A.aeolicus LeuRSs and partial sequence alignment of the four β subunit peptides investigated. Amino acid sequences were aligned in the four peptidic regions here studied. Aa, A.aeolicus; Ec, E.coli; Bs, Bacillus subtilis; MSc, mitochondrial S.cerevisiae; Mt, Mycobacterium tuberculosis; Dr, Deinococcus radiodurans; Uu, Ureaplasma urealyticum; Cp, Chlamydia pneumoniae AR39; Cm, Chlamydia muridarum; Hp, Helicobacter pylori; Hi, Haemophilus influenzae; Mp, Mycoplasma pneumoniae; Sc, Streptomyces coelicolor; Nm, Neisseria meningitidis; Tt, T.thermophilus. Amino acids that are completely conserved in all sequences are boxed in black. Homologous residues are boxed in gray. The residues studied in this work with the yeast 3HS are marked with an arrow. The indicated secondary structure elements are from the T.thermophilus LeuRS 3D structure (7) and from the prediction of the GCG SeqWeb 2.0 software for the C-terminal domain.
