Fig. 1.
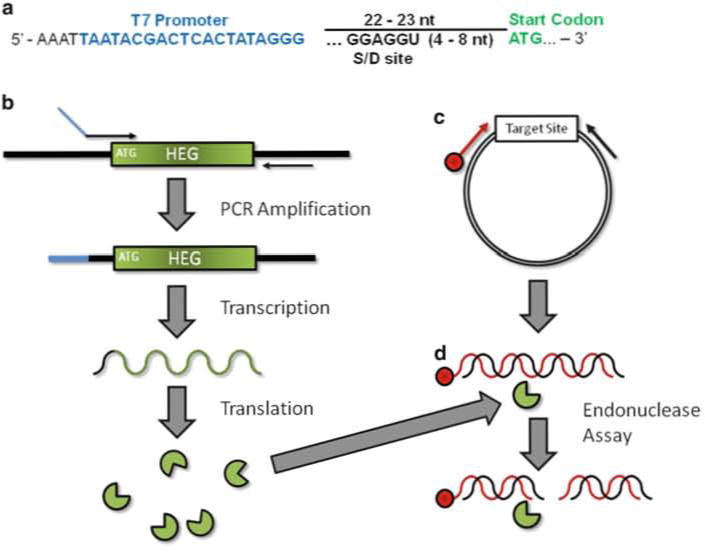
Schematic of in vitro homing endonuclease expression and endonuclease assay. (a) Upstream primer design for amplification of a HEG. T7 promoter (blue) should be placed ~22–23 bp upstream of the HEG initiation codon (green). Sequence at the 5′ end in black is thought to stabilize RNAP–promoter interaction. If using a prokaryotic system a ribosome binding site/Shine–Delgarno sequence (S-D) should be incorporated. (b) In vitro homing endonuclease production. PCR amplification results in the incorporation of a T7 promoter (blue) upstream of the HEG. This product is used to direct protein synthesis in the coupled in vitro transcription/translation reaction. (c) Target site DNA is amplified from a plasmid with a 5′-end labeled (red) and unlabeled primer resulting in a duplex DNA molecule labeled on one strand. The same labeled primer and plasmid DNA is used to generate a DNA sequencing ladder. The resulting ladder and the PCR product are labeled at the exact same position. (d) Endonuclease Assay. The HE (from b) is mixed with precursor dsDNA labeled on the 5′-end. Cleavage of both strands results in two dsDNA products. Of the resulting four individual strands, only one is covalently linked to the label and therefore visible by phosphor imaging
