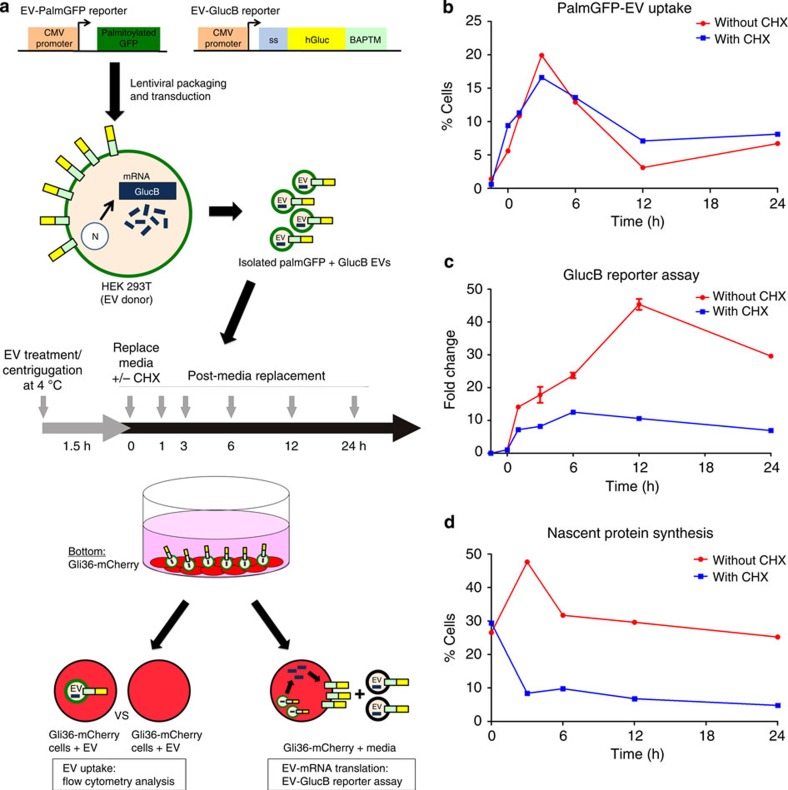
Figure 6. EV uptake and EV-RNA translation.
(a) Schematic of two expression reporters used to detect EV uptake (PalmGFP) and translation of EV-delivered GlucB mRNA (luciferase activity). 293T cells (donors) were stably transduced with lentivirus vectors expressing these reporters. EVs were isolated from conditioned medium of these cells and added to Gli36-mCherry glioma cells followed by centrifugation at 4 °C to facilitate EV docking on cells while minimizing EV uptake. PalmGFP+GlucB+ EV-containing media was then replaced with EV-depleted media in the presence and absence of cycloheximide (CHX) and cells were incubated at 37 °C for 24 h. Medium and cell samples were collected at indicated time points to assess EV uptake (fluorescence and luciferase activity) and EV-mRNA translation (luciferase activity). CMV, cytomegalovirus enhancer/promoter; ss, signal sequence; BAPTM, biotin acceptor peptide with transmembrane domain of platelet-derived growth factor receptor. (b) Flow cytometry analysis of the percentage of Gli36-mCherry cells positive for PalmGFP+ EV uptake in the presence or absence of CHX. Cells that have taken up EVs exhibited signal for both PalmGFP (EVs) and mCherry (recipient cells), whereas cells without or with low levels of EVs showed only mCherry (see also Supplementary Fig. 5). (c) GlucB reporter assay was performed to detect translation of EV-delivered GlucB mRNA by Gli36-mCherry cells expressed as fold change. (d) Flow cytometry analysis of nascent protein synthesis in Gli36-mCherry cells revealed by Click-iT HPG labelling and Alexa Fluor488 conjugation expressed as percentage of cells positive for Alexa Fluor 488 (see also Supplementary Fig. 6).