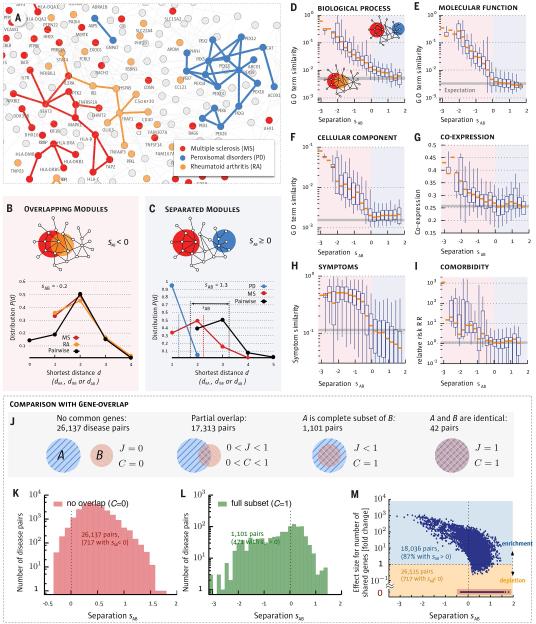
Fig. 3. Network Separation and Disease Similarity.
a, A subnetwork of the full interactome highlighting the network-based relationship between disease genes associated with three diseases identified in the legend. b,c, Distance distributions for disease pairs that have topologically overlapping modules (sAB < 0, b) or topologically separated modules (sAB > 0, c). The plots show P(d) for the disease pairs shown in (a). d-i, Topological separation vs. biomedical similarity. d,e,f, GO term similarity; g, gene co-expression; h, symptom similarity for all disease pairs in function of their topological separation sAB. We highlight in red the region of overlapping disease pairs (sAB < 0) and in blue the separated disease pairs (sAB > 0). For symptom similarity we show the Cosine similarity (cAB = 0 if there are no shared symptoms between diseases A and B and cAB = 1 for diseases with identical symptoms). Comorbidity in (i) is measured by the relative risk RR (40). Bars in d-i indicate random expectation (SM Sect. 1): in d-g the expected value for a randomly chosen protein pair is shown. In h-i the mean value of all disease pairs is used. j-m, The interplay between gene-set overlap and the network- based relationships between disease pairs. j, The relationship between gene-sets A and B is captured by the overlap coefficient C =|A∩B|/min(|A|,|B|) and the Jaccard-index J=|A∩B|/|A∪ B|. More than half (59%) of the disease pairs do not share genes (J = C = 0), hence, their relation cannot be uncovered based on shared genes. k, Distribution of sAB for disease pairs with no gene-overlap. We find that despite having disjoint gene sets, 717 diseases pairs have overlapping modules (sAB < 0). l, Distribution of sAB for disease pairs with complete gene-overlap (C = 1) shows a broad range of network-based relationships, including non-overlapping modules (sAB > 0). m, Fold-change (fc) of the number of shared genes compared to random expectation vs. sAB for all disease pairs. The 59% of all disease pairs without shared genes are highlighted with red back- ground. For 98% of all disease pairs that share at least one gene the gene-based overlap is larger than expected by chance. Despite this fact most (87%) of these disease pairs are separated in the network (sAB > 0). Conversely, a considerable number of pairs (717) without shared genes exhibit detectable network overlap (sAB < 0).