Fig. 1.
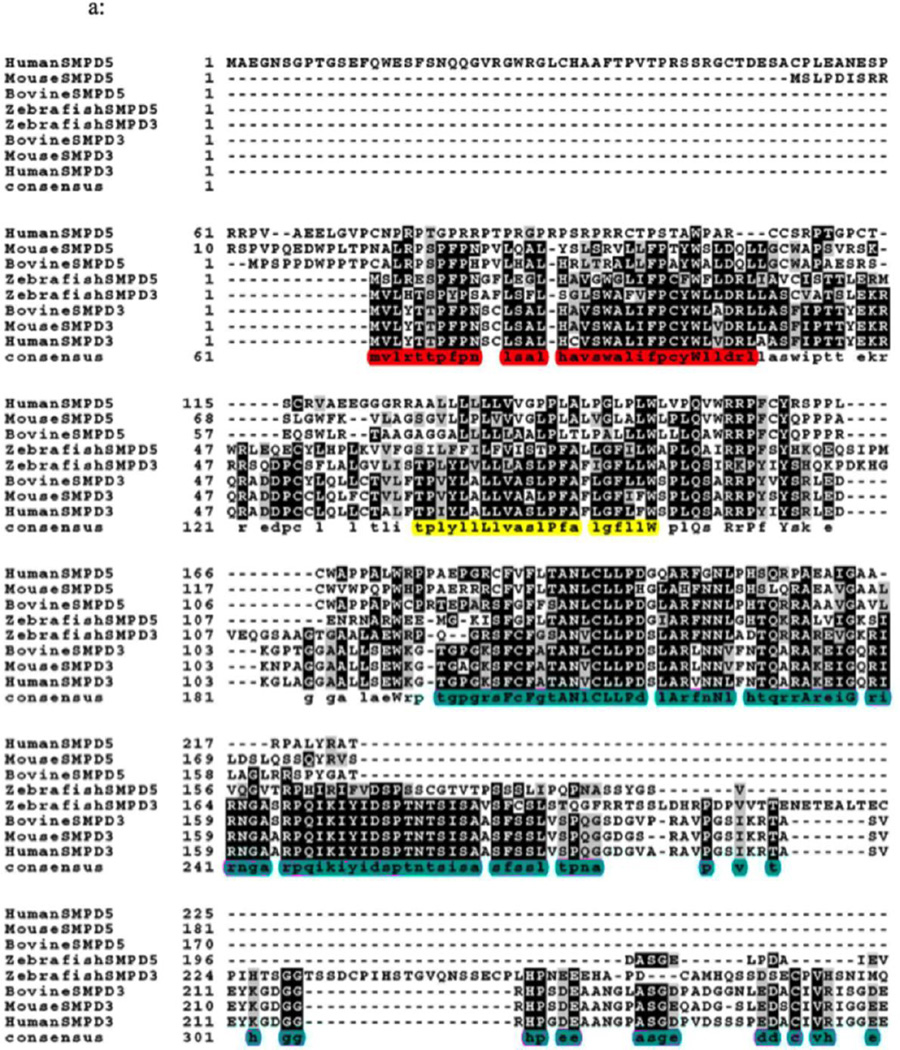
Fig. 1A. Sequence alignment of SMPD 5 (MA-nSMase) with SMPD 3 (nSMase 2). Alignment of the deduced amino acid sequences of human (NP001182466.1), mouse (XP002692632.1), bovine (XP003120137.2) and zebrafish (NP001071083.1) SMPD5; and human (NP001116222.1), mouse (XP005256088.1), bovine (NP001179292.1), and zebrafish (AAH43077.1) SMPD3. The sequences were aligned by Clustal omega program. Identical residues in all the three sequences are indicated by bold characters. The mitochondrial localization signal (MLS) of the zebrafish mitochondrial SMase that spans 1–35aa which is homologous to residues 24–56aa of MA-nSMase highlighted in red, predicted transmembrane domain (TMD) sequence spanning between 77–99aa in MA-nSMase is highlighted in yellow and the catalytic domain that spans 129–483 in the mouse MA-nSMase, predicted based on nSMase 2 sequence is highlighted in pink.
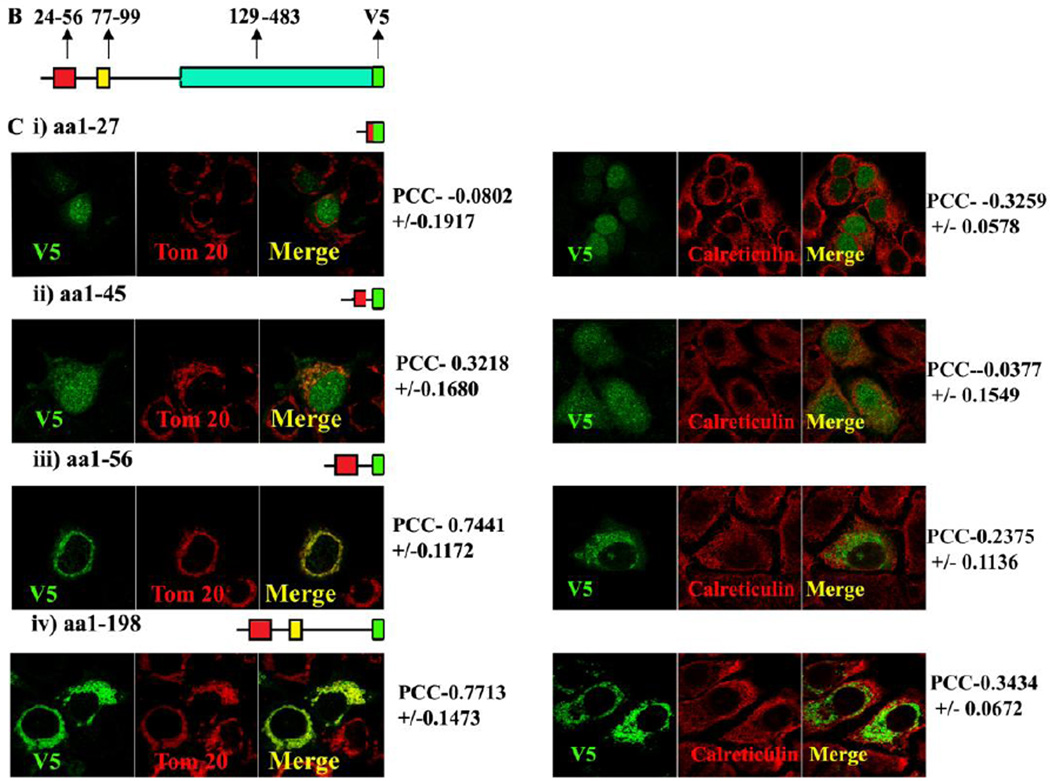
Fig. 1B, Schematic representation of the MLS, TMD and catalytic domain of MA-nSMase. C, Truncated mutant constructs and its co-localization with mitochondrial marker (Tom 20). Table 1 showing the summary of the results of truncation mutant constructs.