FIG 7 .
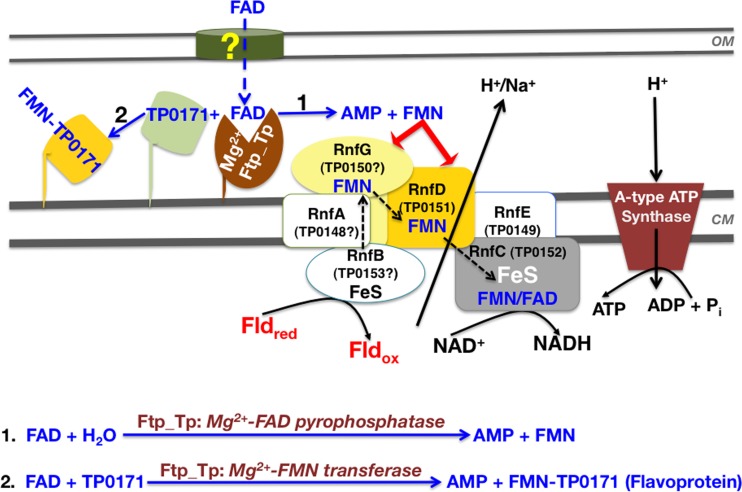
Proposed model of periplasmic flavoprotein biogenesis and flavin homeostasis in the T. pallidum periplasm. The model is predicated on Ftp_Tp’s dual function in posttranslational protein flavinylation described herein and flavin homeostasis modulated by Ftp_Tp’s Mg2+-FAD pyrophosphatase activity (16). The yellow question mark indicates the unknown import mechanism of host-derived FAD across the outer membrane. FAD in the T. pallidum periplasm is either hydrolyzed into AMP and FMN to maintain the flavin pool (scheme 1) and/or utilized by Ftp_Tp’s Mg2+-FMN transferase activity to generate flavoproteins (e.g., TP0171) (scheme 2). The product inhibition of Ftp_Tp’s FAD pyrophosphatase activity (16) likely maintains the FAD pool for FMN transferase activity. The scheme also proposes a hypothetical energy conservation pathway that likely couples a noncanonical flavin-based Rnf redox pump to T. pallidum’s A-type ATP synthase (see text for explanation). Note that the function of the redox protein(s) depends on periplasmic flavin and the dual activities of Ftp_Tp. Assignments of the unidentified Rnf subunits (designated by “?”) ostensibly encoding the T. pallidum Rnf complex (the tp0153-tp0148 operon) are based tentatively on an rnfBCDGEA type of system (24) and the predicted transmembrane helices of TP0148, TP0149, and TP0150. Also, TP0149 has ~30% identity to Pseudomonas brassicacearum RnfE (according to the KEGG gene function identification tool). OM, outer membrane; CM, cytoplasmic membrane; ox, oxidized; red, reduced.