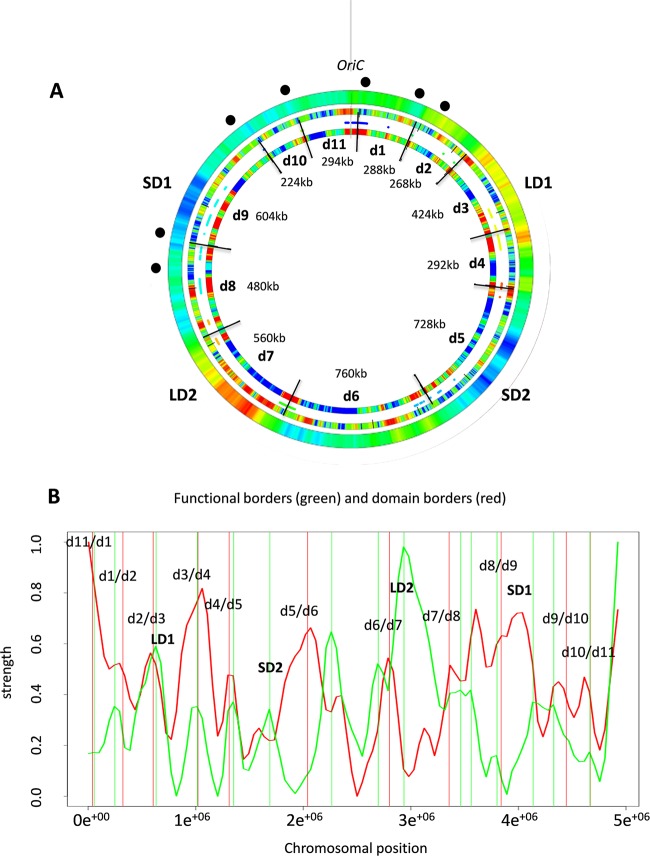
FIG 3 .
Stress-response domains. (A) Compilation of the spatial boundaries based on pairwise correlations of different DNA sequence parameters in D. dadantii genome. The outermost ring shows the average negative melting energy distributions in the genome. The locations of rrn genes are marked by black dots. The middle ring represents the static GO tree functional boundaries. Red indicates a substantial change in the functional composition of genes in the genome (35). The innermost ring shows the detected 11 stress-response boundaries delimiting 11 domains (d1 to d11). Red indicates the statistically significant change in the expressed DNA sequence parameter values. (B) Comparison of the functional borders (green vertical lines) and domain borders (red vertical lines). The change of function (green curve) and the correlation of domain borders in the expression profiles (red curve) are depicted in arbitrary units. The coincidence of both borders was tested by a random model. Only 2.4% of randomly chosen border positions show a smaller median nearest-neighbor distance between both sets of borders.