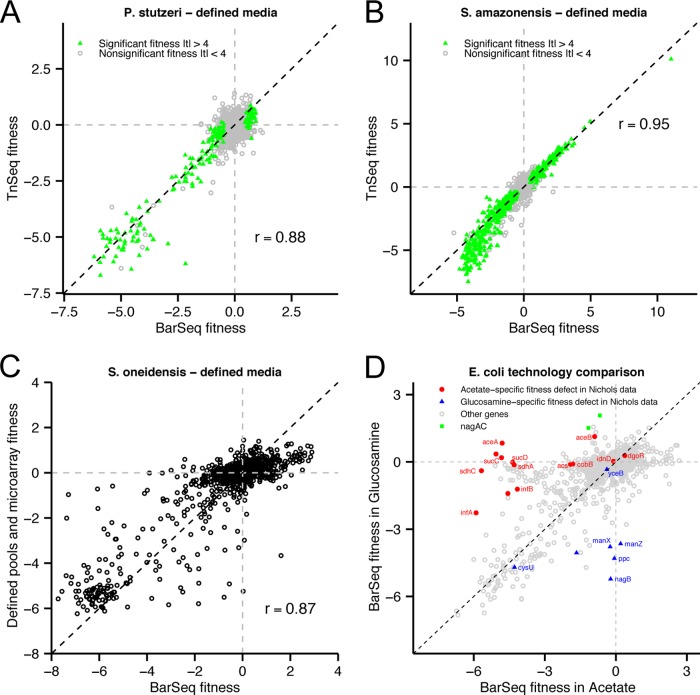
FIG 3 .
Comparison of RB-TnSeq to other technologies. (A) Comparison of gene fitness for P. stutzeri grown in a defined medium with glucose as determined with BarSeq (x axis) or sequencing the transposon-genome insertion junctions (TnSeq; y axis), starting from the same samples of genomic DNA. Genes marked in green have statistically significant phenotypes as determined by BarSeq. The dashed black line marks x = y. (B) Same as panel A for S. amazonensis grown in a defined medium with d,l-lactate. (C) Comparison of S. oneidensis gene fitness in defined medium with l-lactate calculated from BarSeq (x axis) and previously described data that used mutant libraries with defined DNA bar codes and microarrays to assay strain abundance (y axis) (2). The dashed black line marks x = y. (D) BarSeq fitness data for E. coli genes grown in acetate (x axis) or glucosamine (y axis) as the sole source of carbon. Genes marked in red have an acetate-specific fitness defect while those marked in blue have a glucosamine-specific fitness defect in the Nichols et al. data set, with thresholds of S < −5 and S > −2 (4).