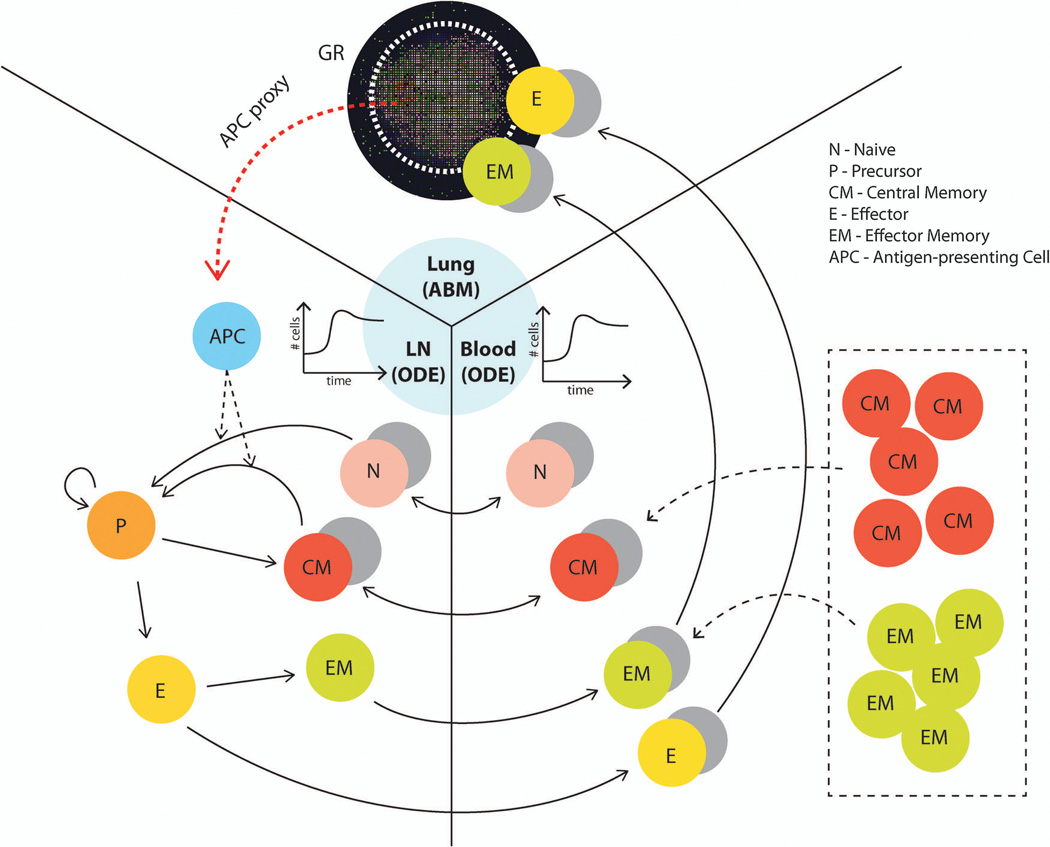
Figure 10.
Three-compartment model framework for simulating the influence of memory cells (which can be generated by vaccines) on granuloma formation and function. The lung (site of infection) is represented with our agent-based model (GranSim) as described in previous sections, and two ODE models capture LN and blood dynamics. T cells that are tracked in LNs include: CD4+ and CD8+ T cells, and each of these can be further classified into: N (Naïve), CM (Central Memory), EM (Effector Memory), P(precursor cells), E (Effector). APCs such as DCs circulate from the lung to the LN to prime the adaptive immune response. The CM, EM, E and N classes can travel between LN and blood compartments as indicated by arrows. Only E and EM (total effector class) can travel to the infection site in the lung. Both M. tuberculosis-specific and non-specific T cells are accounted for in our model. For our in silico experiments, we changed the initial conditions of equations describing the number of different memory cells in the blood to represent the memory cells that we assume have been generated after vaccination (shown in box). The cell and bacterial time courses and granuloma spatial outcomes in the lung are tracked to assess the level of protection derived from the simulated vaccine.