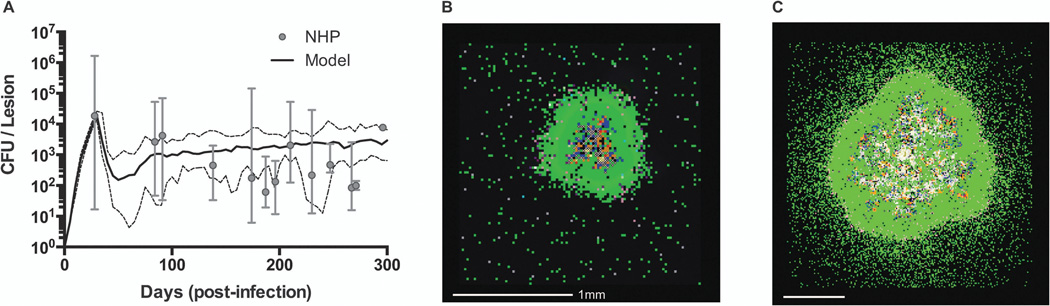
Figure 4.
Granuloma model calibration and snapshots. (A) Comparison of CFU/lesion data from non-human primates (NHP) with computational model predictions (median – solid black line, min/max – dashed black lines). Model detail and parameters are given in [52]. NHP data from 32 animals collected between 28 and 296 days post-infection has been previously published in[10, 17]. (B) Sample simulation snapshot shows a granuloma that is containing infection at 200 days post-infection. (C) Sample simulation snapshot with different parameter values than in (B) shows a granuloma that fails to contain infection. Snapshot legend colors: resting macrophages (green), infected macrophages (orange), chronically infected macrophage (red), activated macrophage (dark blue), pro-inflammatory T cell (pink), cytotoxic T cell (purple), regulatory T cell (aqua), extracellular bacteria (brown), and caseation (cross-hatch).