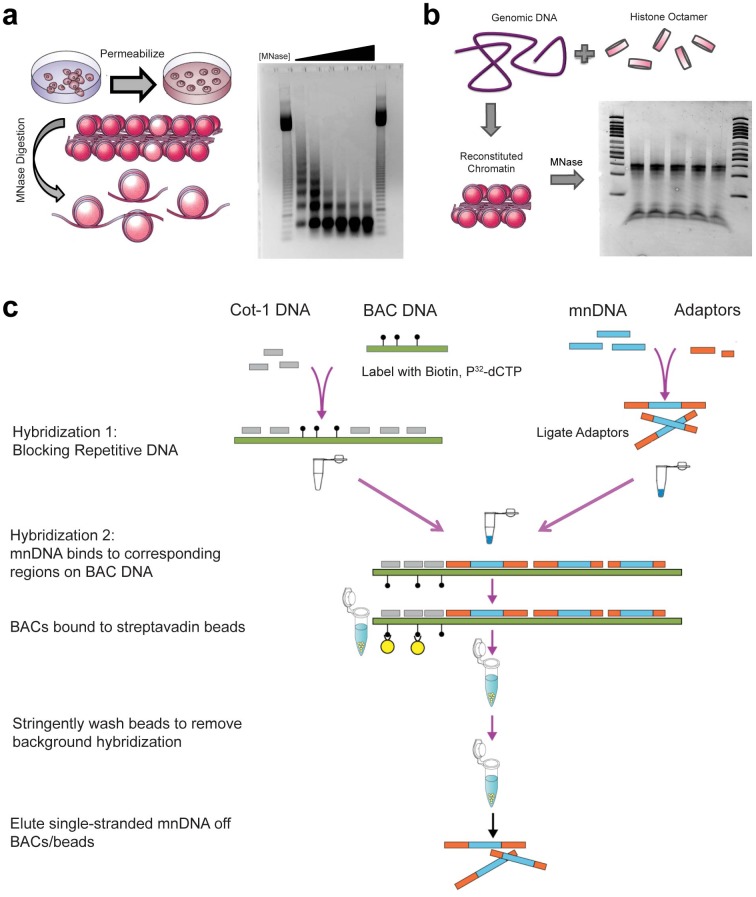
Fig 1. Mapping in vivo and in vitro nucleosome occupancy using BAC-based enrichment.
(a) Protocols were modified so that permeabilization and micrococcal nuclease digestion occur while embryonic stem cells are attached to the tissue culture surface to improve recovery and digestion reproducibility. The amount of micrococcal nuclease (MNase) in the digestion was titrated so that the mononucleosome band at 147bp is the primary band, without overdigestion. Digests were measured in Worthington Units of MNase * Time of digestion / volume of cell culture (U*min/mL). Lanes 1 and 8: 50bp ladder. Lanes 2–7 range from 2500 U*min/mL to 25,000 U*min/mL of MNase, with ideal digestion in the third condition, 10,000 U*min/mL. This amount of digestion was used in all future experiments. (b) Genomic DNA was purified from embryonic stem cell cultures and combined with histone octamer purified from chicken erythrocytes in a ratio of 100μg:30μg under high salt (2M NaCl) conditions. Removal of salt via dialysis results in reconstituted chromatin, representing histone proteins’ preferred DNA sequences. Reconstituted chromatin was digested with 5 Worthington Units of micrococcal nuclease per 10μg of genomic DNA present, for a digestion of 5 minutes at 37°C (Lanes 1 and 7: 50 bp ladder; Lanes 2–6: digested chromatin). (c) Bacterial Artificial Chromosome (BAC) DNA, which was nicked with biotin-dUTP, was blocked with Cot-1 DNA at a ratio of 100ng:10μg. 1μg of library-adapted mononucleosome DNA was denatured and mixed with BAC DNA. Mononucleosome DNA was hybridized to the corresponding BAC region and was isolated by removing BACs from solution with streptavadin beads, stringently washing the beads, and eluting single stranded DNA from the beads. Double stranded products were amplified using PCR and sent for paired-end sequencing.