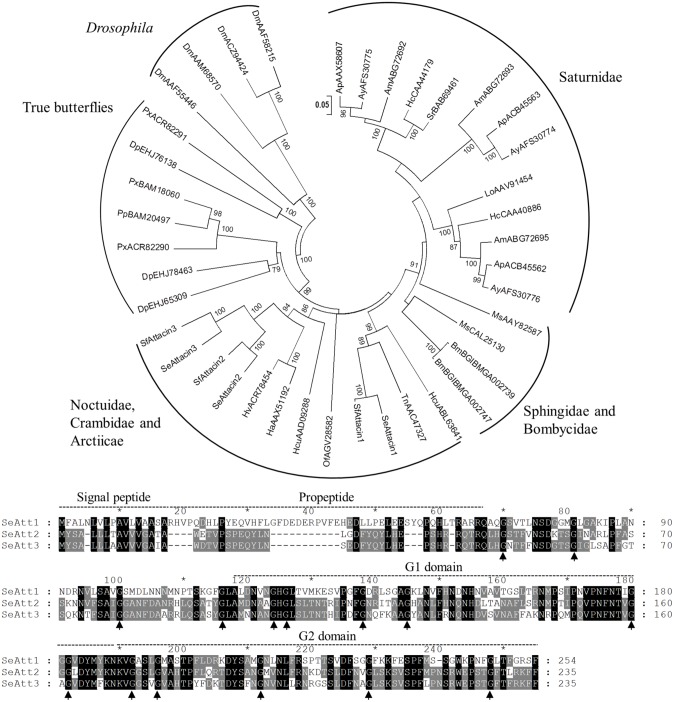
Fig 1. Phylogenetic analysis of attacin proteins from Lepidoptera.
Upper panel) Neighbor-joining tree of Lepidoptera attacins. D. melanogaster attacins were used as outgroup. Bootstrap values (above 75) are indicated on each node of the tree (100 replicates). Sequences from nr database, SPODOBASE and Silkworm Genome Database were used. Accession number of each sequence is indicated. Abbreviations: Am: A. mylitta, Ap: Antheraea pernyi, Ay: Antheraea yamamai, Bm: B. mori, Dm: D. melanogaster; Dp: D. plexippus, Ha: H. armigera, Hv: H. virescens, Hc: H. cecropia, Hcu: Hyphantria cunea, Lo: Lonomia obliqua, Ms: Manduca sexta, Of: O. furnacalis, Pp: Papilio polytes, Px: Papilio xuthus, Sc: Samia cynthia, Se: S. exigua, Sf: S. frugiperda, Tn: T. ni. Lower panel) ClustalX alignment of SeAttacins. Black shadows highlight residues conserved between the three isoforms whereas grey shadows highlight residues conserved among two isoforms. Arrows indicate conserved glycine residues.