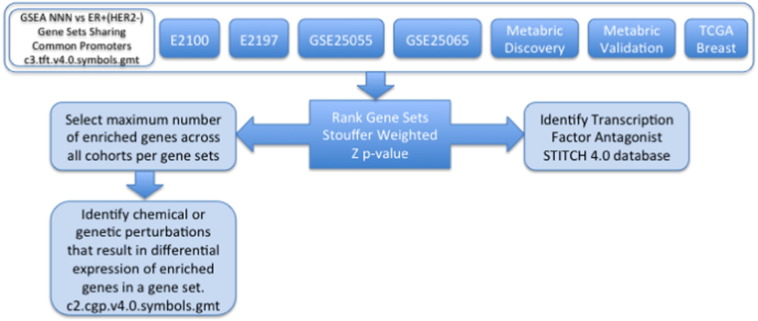
Fig. 1.
Each cohort consisting of TN and ER+(HER2 −) samples are run using GSEA to determine gene sets that are enriched and share a common promoter motif. The p-value from each enriched gene set is combined and ranked using Stouffer weighted Z to identify gene sets that have consistent enriched gene sets across all cohorts. The transcription factor for each ranked enriched gene set are searched in the STICH 4.0 database for chemical inhibitors or activators. Additionally, common set of genes in each gene set shown to be enriched across maximum number of cohorts are searched for known chemical and genomic perturbation gene set to identify possible inhibitors or activators.