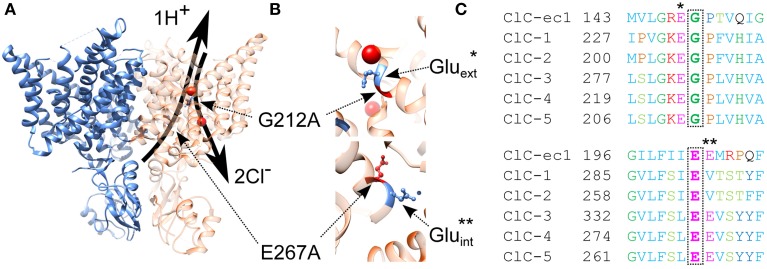
Figure 1.
Structural localization and evolutionary conservation of two Dent's disease associated mutations. (A) Ribbon representation of the dimeric biological assembly typical for proteins of the CLC family and based on the crystal structure of the eukaryotic CmCLC (PDB id: 3ORG). Two identical monomers capable of mediating coupled chloride/proton exchange are assembled to form a functional CLC dimer. Superimposed as arrows on the right monomer are the anion and proton transport pathways, separated at the bottom (intracellular) side and converging at the central anion biding site to follow the same exit/entry pathway toward the top of the protein. The two here investigated Dent's disease mutations are localized at key positions along these pathways—mutation G212A is located at the top of the protein at the separate exit for anions and protons; mutation E267A is localized at the intracellular entrance of the proton transport pathway. (B) Enlarged view of the protein regions surrounding mutations G212A and E267A with additionally annotated the gating [Gluext – indicated with a star here and in the sequence alignment in (C)] and proton [Gluin – indicated with two stars here and in the sequence alignment in (C)] glutamates that both play decisive roles for the CLC transport cycle. (C) Sequence conservation of the regions surrounding both investigated Dent's disease mutations. The glycine G212 is an amino acid that is conserved throughout the mammalian CLC isoforms and precedes the gating glutamate Gluext. Glutamate E267 is also well-conserved and precedes the proton glutamate E268 in the sequence of ClC-5.