FIGURE 5.
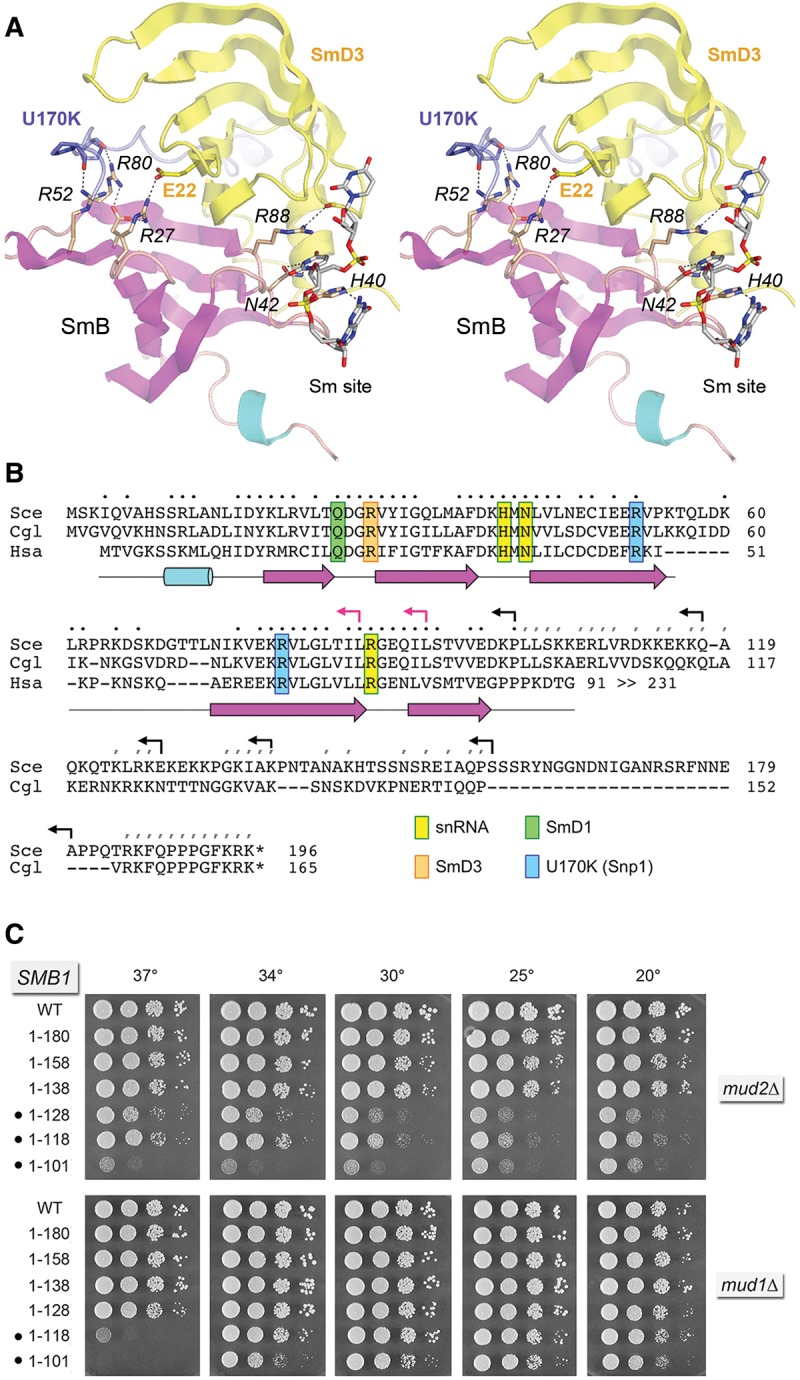
Structure-guided mutagenesis of SmB. (A) Stereo view of the human U1 snRNP structure highlighting the fold of SmB (depicted as a cartoon trace with magenta β strands and cyan helices) and its interactions with neighboring subunits SmD3 (yellow) and U1-70K/Snp1 (blue) and with the Sm site in U1 snRNA. Selected amino acids are shown as stick models and numbered according to their positions in the yeast polypeptides. Atomic contacts are indicated by dashed lines. (B) Alignment of the primary structures of the S. cerevisiae (Sce), Candida glabrata (Cgl), and human (Hsa) SmB. Positions of side-chain identity/similarity in the amino-terminal segments of all three proteins are indicated by • above the alignment. Positions of side chain identity/similarity between the carboxy-terminal segments of SceSmB and CglSmB are denoted by /. The secondary structure elements of the amino-terminal domain are depicted below the alignment, with β strands as magenta arrows and α helices as cyan cylinders. SmB amino acids that make contacts to other U1 snRNP subunits and contact the U1 snRNA are highlighted in color-coded boxes as indicated. Reverse arrowheads indicate the boundaries of the carboxy-terminal truncations of yeast SmB; black and red arrowheads denote viable and lethal truncations, respectively. (C) The wild-type and truncated SMB1 alleles were tested for activity by plasmid shuffle in smb1▵ mud2▵ and smb1▵ mud1▵ strains. The growth phenotypes of viable FOA-resistant smb1▵ strains bearing the indicated SMB1 alleles are shown. Synthetic defects are denoted by •.
