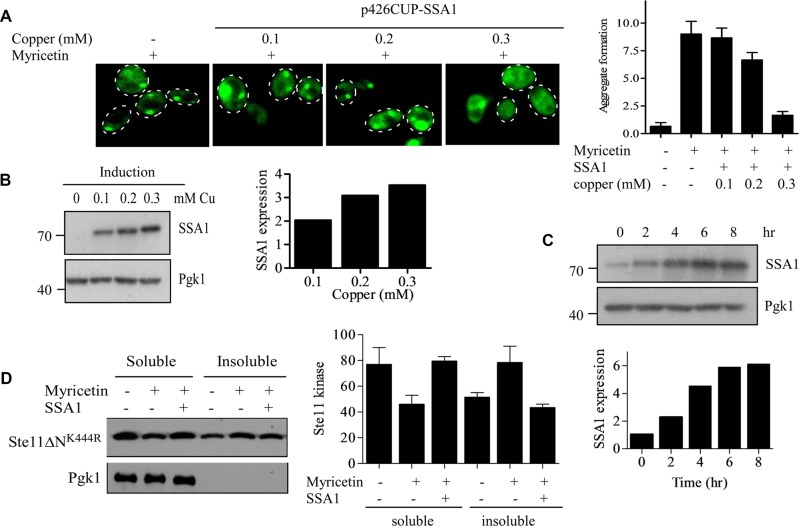
FIGURE 7:
Hsp70 expression from inducible CUP promoter clears kinase inclusions. (A) p426CUP-SSA1 plasmid was transformed into GFP-Ste11ΔNK444R–containing yeast cells. GFP-Ste11ΔNK444R was expressed from the GAL promoter by the addition of galactose. The cells were treated with 100 μM myricetin for 2 h to form the kinase inclusions. SSA1 was then expressed from the CUP promoter for an additional 6 h with varying concentrations of copper ion, as indicated. The GFP fluorescence was observed, and the inclusions were counted in a minimum of three different fields with an average of five cells per field. The quantification of inclusions is shown as a bar diagram. (B) Western blots of cell lysates expressing SSA1 with different concentrations of copper ion. Pgk1 served as a loading control. The bands were quantified by ImageJ software and plotted as a bar diagram. (C) p426CUP-SSA1–containing yeast cells were induced with 300 μM of copper ion for different times, as indicated. The expression of SSA1 was measured by Western blot analysis with anti-His antibody. The bands were quantified, normalized with Pgk1, and plotted as a bar diagram. (D) WT cells were cotransformed with p416GPD-Ste11ΔNK444R (marker-swapped Leu version) and p426CUP-SSA1 plasmids. The cells were treated with 100 μM myricetin for 2 h. The SSA1 was expressed by the addition of 300 μm of copper ion for 6 h. The soluble and insoluble fractions were separated and analyzed by Western blotting. The bands were quantified and plotted as a bar diagram. Bar represents SE of three independent experiments.