FIGURE 1:
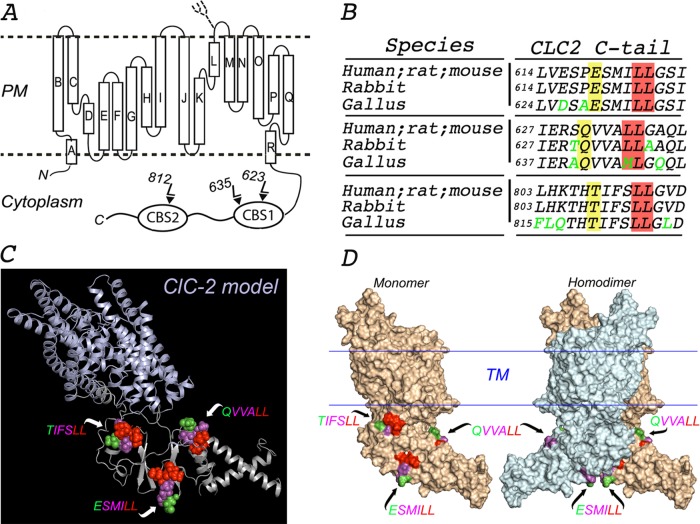
In silico molecular model of ClC-2. (A) Proposed topology of human ClC-2. ClC-2 displays 18 α-helices (A–R) that span the membrane, both N- and C-terminus ends facing the cytoplasm. The C-terminus of ClC-2 contains the CBS1 and CBS2 domains, which display two and one dileucine motifs, respectively. A single N-linked glycan is present on the short luminal loop between the L and M α-helices, as indicated. (B) Multiple sequence alignment using Clustal Omega program (UniProt) with sequences from the indicated species (P51788, human; P35525, rat; Q9R0A1, mouse; P51789, rabbit; XP 423073.4, gallus). The amino acid sequences around each of the three dileucine motifs in the cytoplasmic tail of ClC-2 are conserved in mammals (human, rat, mouse, and rabbit). One of the two leucines in the QVVA645LL motif is changed to a methionine (QVVA645ML) in birds (gallus). Conserved residues are shown in black, and nonconserved residues are shown in green. The critical leucine pair and residues at position −4 are shaded in red and yellow, respectively. (C) Combined cartoon-rendered structure in a monomer of ClC-2 displaying the three dileucine motifs in a sphere format. We modeled the structure of ClC-2 (UniProt P51788) using MODELLER and available crystal structure information for the transmembrane and extracellular domains of CmClC (3ORG), EcClC (1OTS), and StCLC (1KPL) and for the CBS domains of ClC-Ka (2PFI) and ClC-0 (2D4Z). (D) Surface-rendered structure of ClC-2 monomer (left) and homodimer (right) displaying the three dileucine motifs. TIFS 812LL is present in a region of CBS2 that participates in the dimerization of the channel and is thus hidden in the dimer. For each motif, essential L residues are represented in red, nonessential variable residues at −1, −2, and −3 positions are displayed in magenta, and the residues at the −4 position are shown in green.