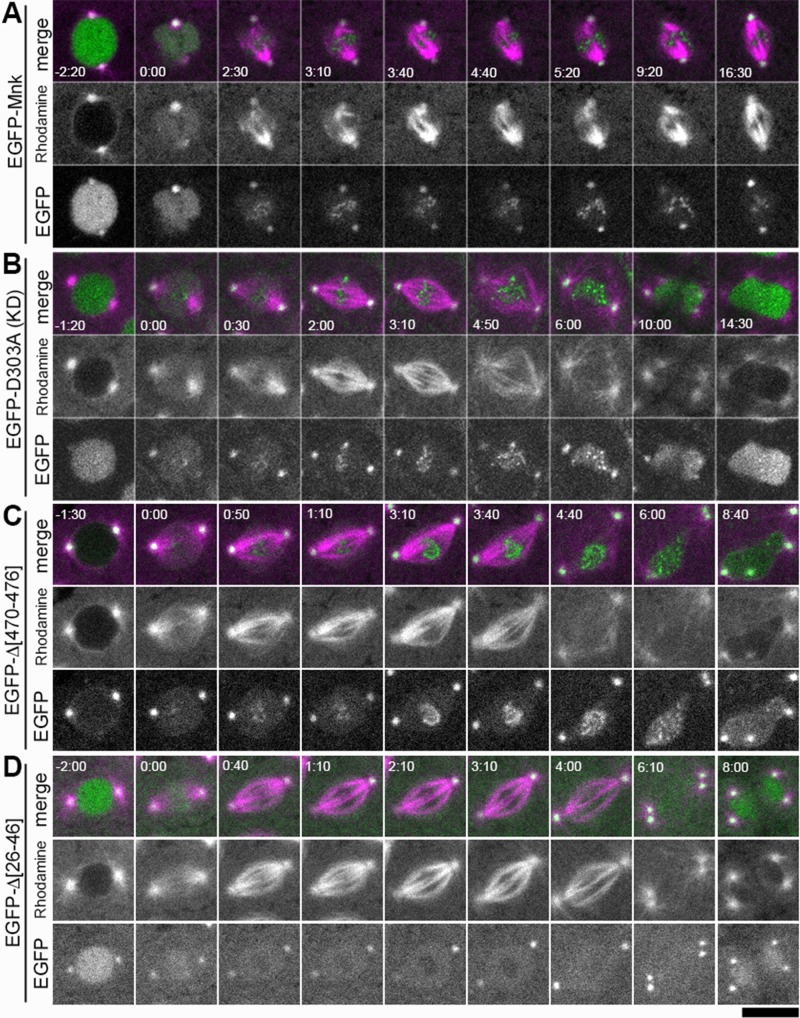
FIGURE 5:
Localization of EGFP-Mnk and Mnk variants with DNA damage. Bleomycin and rhodamine-tubulin were injected into transgenic embryos expressing each EGFP-Mnk variant in mnk mutant background. EGFP is shown in green and rhodamine is shown in magenta in merged images. (A) EGFP-Mnk restored centrosome inactivation and mitotic delay in mnk mutant. Anastral spindles formed (time 2:30–9:20). EGFP-Mnk localized to centrosomes and formed foci/aggregates on mitotic chromosomes (time 2:30–16:30). Frames were selected from Supplemental Movie S5. (B) EGFP-D303A did not restore centrosome inactivation and mitotic delay in mnk mutant. Normal spindles formed (time 2:00–3:10). Mitotic delay did not occur; however, chromosome segregation failed, and connected dumbbell-shaped nuclei formed. The midbody did not form (time 10:00–14:30). EGFP-D303A localized to centrosomes and formed foci/aggregates on mitotic chromosomes (time 0:30–6:00) similarly to EGFP-Mnk. Frames were selected from Supplemental Movie S6. (C) EGFP-Δ[470-476] did not restore centrosome inactivation and mitotic delay in this mnk mutant embryo. EGFP-Δ[470-476] was not actively imported into the nucleus. It localized to centrosomes and formed foci/aggregates on mitotic chromosomes (time 0:50–4:40) similarly to EGFP-Mnk or EGFP-D303A. Frames were selected from Supplemental Movie S16. (D) EGFP-Δ[26-46] did not restore centrosome inactivation and mitotic delay in mnk mutant. EGFP-Δ[26-46] localized to the nucleus and centrosomes. It was excluded from the vicinity of mitotic chromosomes (time 0:40–4:00) as shown without DNA damage (Figure 4E). It did not make foci/aggregates on mitotic chromosomes. Frames were selected from Supplemental Movie S14. Elapsed time is shown in minutes:seconds. NEB is set as time 0:00. Scale bar, 10 μm.