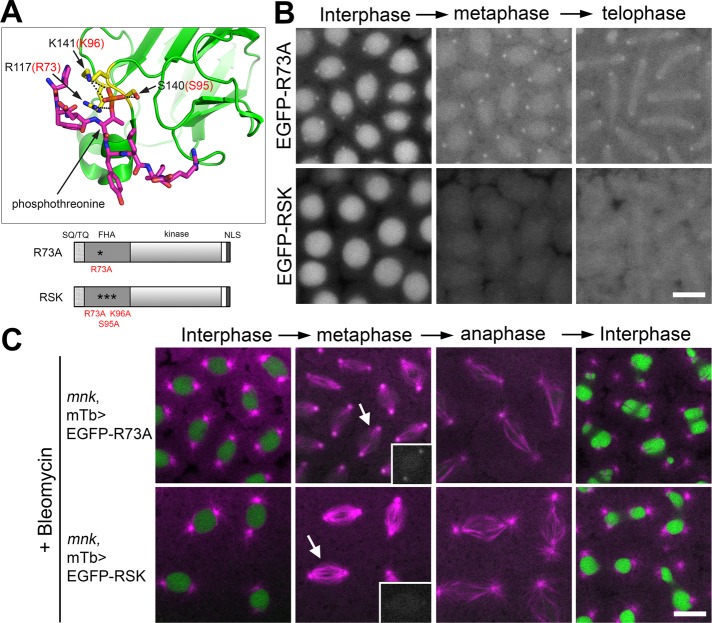
FIGURE 7:
The phosphopeptide-binding ability of the FHA domain is required for Mnk localization to key mitotic structures. (A) The 3D structure of part of the human Chk2 FHA domain (green) and a synthetic phosphopeptide HFDpTYLI (magenta) based on an x-ray structural study by Li et al. (2002; Protein Data Bank ID: 1GXC). Prepared using PyMOL (www.pymol.org). The phosphate of pThr is shown in orange. Three conserved residues (R117, S140, and K141) that form hydrogen bonds (black dotted lines) with the phosphate are shown in yellow. Corresponding amino acid numbers of the three residues in Drosophila Mnk are shown in red (R73, S95, K96). Schematic diagrams of Mnk variants that carry R73A or RSK mutation are shown below the 3D structure. (B) R73A mutation greatly reduced localization of the molecule to centrosomes, interkinetochores/centromeres, and the midbody and disrupted localization to pseudocleavage furrows. RSK mutation completely disrupted localization of the molecule to all the mitotic structures. Frames were selected from Supplemental Movies S17 (EGFP-R73A) and S18 (EGFP-RSK). Scale bar, 10 μm. (C) EGFP-R73A and EGFP-RSK are not functional to restore centrosome inactivation, mitotic delay, and nuclear dropping responses after bleomycin injection in mnk mutant embryos. EGFP signals are shown in green and rhodamine signals in magenta. Frames were selected from Supplemental Movies S19 (mnk, mTb>EGFP-R73A) and S20 (mnk, mTb>EGFP-RSK). Insets, EGFP signals on spindles indicated by arrows. Scale bar,10 μm.