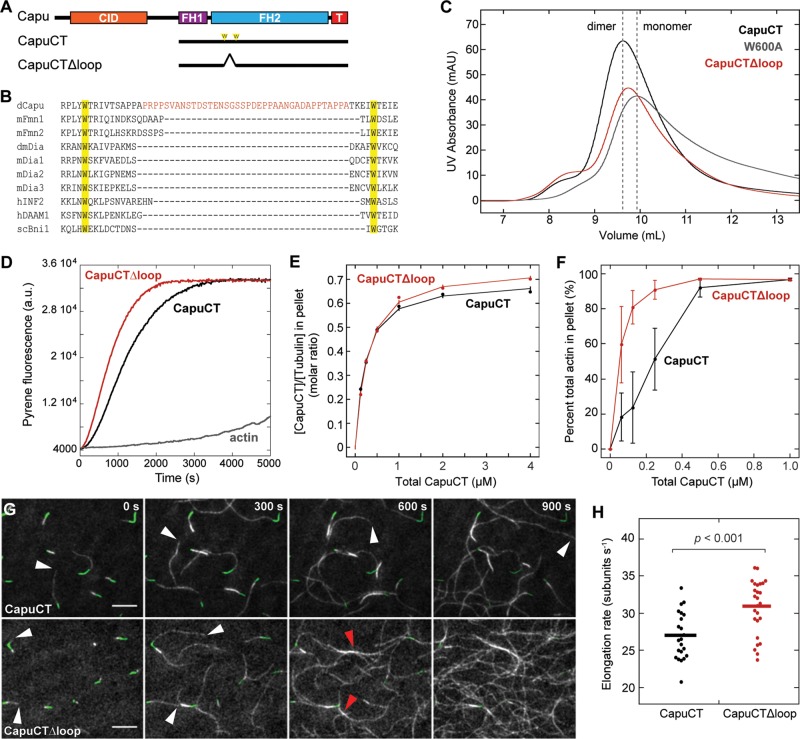
FIGURE 6:
Deletion of a “loop” enhances Capu's actin assembly and bundling activity. (A) Domain organization of Capu (1059 amino acids) and schematics of CapuCT and CapuCTΔloop. (B) Sequence alignment showing the location of Capu's extra “loop” domain between the highly conserved lasso Trp residues. (C) Size exclusion chromatography of CapuCTΔloop compared with wild-type CapuCT (dimer) and CapuCT-W600A (monomer) controls. (D) Actin assembly activity of 10 nM CapuCTΔloop in the presence of 8 μM S. pombe profilin. (E) Microtubule binding by CapuCTΔloop compared with wild type at 50 mM KCl with 0.5 μM tubulin. (F) F-actin bundling by CapuCTΔloop compared with wild type. Each bundling curve represents the mean of at least four independent experiments, with error bars showing the SD. (G) TIRF microscopy observation of actin filament (gray) elongation from immobilized F-actin seeds (green) in the presence of wild-type CapuCT or CapuCTΔloop. White arrowheads denote the barbed ends of fast-growing dim filaments. Red arrowheads points to F-actin bundles. Scale bar, 10 μm. The same conditions were used for all TIRF experiments: 0.6 μM actin (20% Oregon green labeled), 1 nM CapuCT, and 3 μM Chic (Drosophila profilin). (H) Quantification of elongation rates for wild-type CapuCT and CapuCTΔloop. Dots represent individual filaments, and bars represent the mean. The p value was calculated using the Mann–Whitney U test.