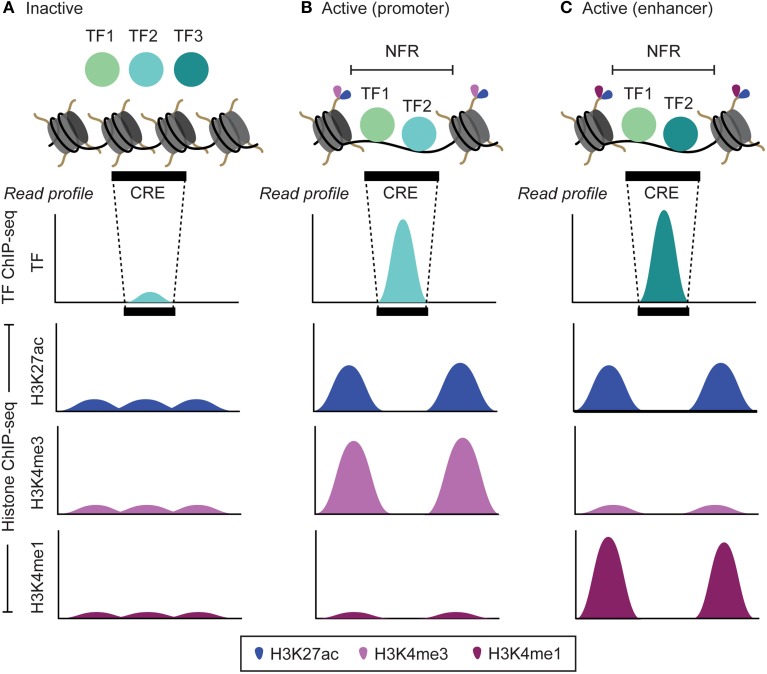
Figure 3.
The transcription factor and histone modification landscape for inactive and active cis-regulatory elements (CRE; promoters and enhancers) and the corresponding read profiles. (A) When inactive, the DNA corresponding to the CRE is wrapped around histone proteins in the form of a basic structural unit termed as nucleosome. This prevents any interaction of transcription factors (TF) with the DNA. (B,C) When active, a series of histone modifications (H3K4me1, H3K4me3, H3K27ac and others) along side interaction with specific TF (pioneer factor) make the overlapping nucleosomes at a CRE hypermobile. These nucleosomes are then displaced apart leading to formation of nucleosome free regions (NFRs) that are subsequently bound by TFs. During TF ChIP-seq and Histone ChIP-seq experiment, reads corresponding to TF bound NFRs and histone-modified regions flanking the NFRs are obtained, respectively. Upon mapping, it leads to a read profile in the form of a peak shape for NFRs (TF ChIP-seq) and peak-valley-peak shape for regions flanking the NFRs (Histone ChIP-seq). By analyzing the read intensity in these read profiles, we can determine active CRE, TF bound at these CRE and also the level of their activity. Since, distinct sets of histone modifications are observed at enhancers (H3K27ac and H3K4me1) and promoters (H3K4me3 and H3K4me1), analyzing peak-valley-peak histone read profile also facilitates to distinguish between enhancers and promoters.