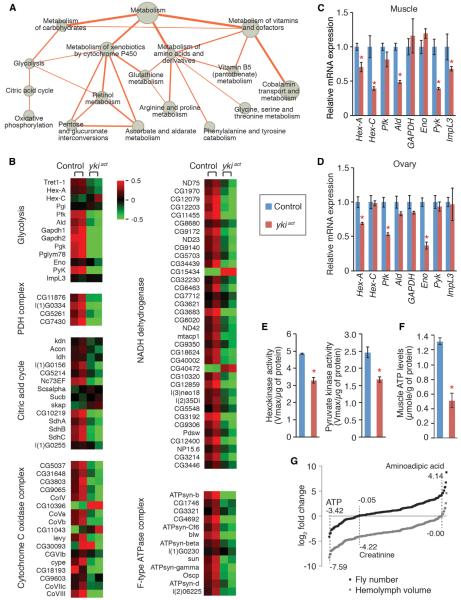
Figure 2. Repression of Energy Metabolism in esgts>ykiact Muscles.
(A) Network presentation of gene list enrichment analysis results focusing on metabolism. All presented metabolic processes are identified to be significantly downregulated inmuscles of esgts>ykiact flies (Tables S2 and S3).Node size indicates enrichment (−log100 p value), and edge thickness represents the number of common genes between two gene sets. The citric acid cycle and pyruvate metabolism (p = 0.0253, GSEA) and oxidative phosphorylation (p = 0.000, GSEA) are identified by GSEA analysis (Subramanian et al., 2005).
(B) Representative downregulated processes and complexes involved in energy metabolism. Heat map signal indicates log2 fold-change values relative to the mean expression level within the group. Red signal denotes higher expression and green signal denotes lower expression relative to the mean expression level within the group. Related GSEA plots are shown in Figure S1.
(C and D) Relative mRNA expression of glycolytic enzyme in muscle (C) and ovaries (D). Measurements shown are mean ± SDs.
(E) Activities of Hexokinase) (left) and Pyruvate kinase (right) in muscle at 8 days of induction (mean ± SEMs).
(F) ATP levels in muscle at 20 days of transgene induction.
(G) Metabolomic analysis of hemolymph metabolites. The metabolite counts are normalized to fly number (black) or extracted hemolymph volume (gray). Log2 fold-change values of the metabolites in hemolymph of esgts>ykiact flies relative to control are presented.
Reduced 55 metabolites in hemolymph of esgts>ykiact flies are shown in Figure S2. *p ≥ 0.05 (Student's t test) compared to control. Genotypes are asshown in Figure 1.