Figure 5.
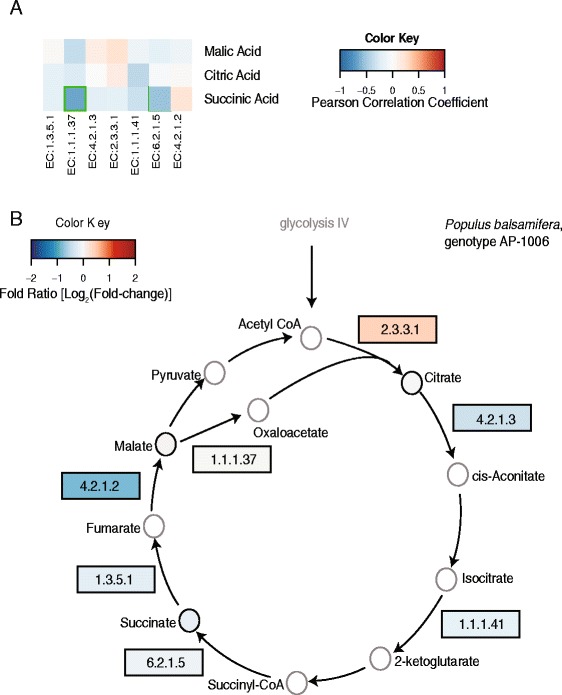
Pathway analysis related to the citric cycle (TCA cycle). (A) Correlation among selected transcripts and metabolites from the KEGG pathway pop00020 ‘Citrate cycle (TCA cycle)’ for genotype AP-1006. Colors represent Pearson correlation value. Red indicates positive correlation and blue represents negative correlation values. (B) Map displays selected steps from citrate cycle pathway. Colours indicate fold-change in transcript or metabolite abundance between water-deficit and well watered treated samples for genotype AP-1006; red indicates higher abundance in water-deficit-treated samples and blue indicates lower abundance in water-deficit-treated samples. Enzymes are given as EC numbers. EC 1.1.1.37, malate dehydrogenase; EC:1.1.1.41, isocitrate dehydrogenase (NAD+); EC:1.3.5.1, succinate dehydrogenase; EC:2.3.3.1, citrate synthase; EC:5.2.1.2, fumarate hydratase, EC: 5.2.1.3, aconitate hydratase, EC: 6.2.1.5, succinate-CoA ligase, beta subunit.
