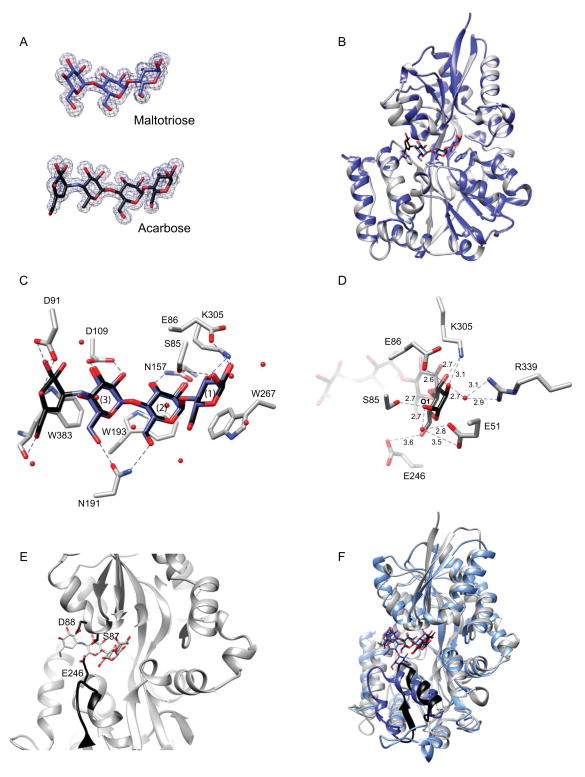
Figure 5. Structure of the maltodextrin-binding protein EUR_01830.
A. Omit maps (σ = 3.0) displaying representative electron density for maltotriose and acarbose. B. Overlay of the structures of EUR_01830 complexed with acarbose (grey ribbon) and with maltotriose (blue ribbon). Acarbose is displayed as black sticks and maltotriose as blue sticks. The aromatic residues that cradle the maltodextrins are shown as sticks. C. Close-up of the binding pocket of EUR_01830. Direct hydrogen-bonding interactions between the protein and maltrotriose/acarbose are displayed as dashed lines. K91 and D384 have been omitted for clarity. Waters involved in water-mediated hydrogen-bonds are shown as spheres. D. Close-up end view of the hydrogen-bonding network that confers recognition of the reducing end sugar. Acarbose and the relevant amino acids are shown as sticks, while waters involved in water-mediated hydrogen bonds are shown as red spheres. Dashed lines indicate interactions within hydrogen bonding distance, and distances are displayed in Å. Here E246, though unlikely to contribute to hydrogen-bonding, is shown for reference. E. Close-up side view of EUR_01830 binding pocket demonstrating the manner in which E246, S87 and D88 may occlude the front of the binding pocket. The loop of residues 242–248 is shown in black ribbon, and E246, S87 and D88 are shown in black sticks. Note that S87 was observed in two conformations in the crystal structure. F. Superposition of the coordinates of the acarbose-bound EUR_01830 (grey) and MalX of S. pneumoniae (blue, pdb 2XD3). The C-terminal strand-loop-helix comprising residues 236–259 of EUR_01830 is displayed in black, and the corresponding helix-loop-helix comprising residues 240–266 of MalX is displayed in dark blue. Acarbose is displayed as black sticks and maltoheptaose is displayed as blue sticks.