Figure 3.
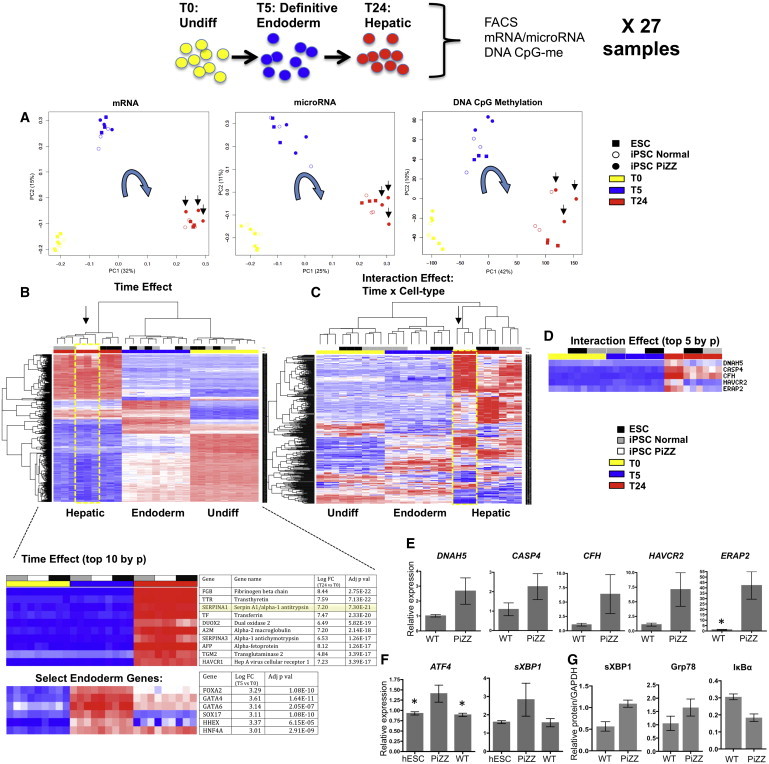
Genome-wide Transcriptomic and Epigenomic Analysis of PSCs at Key Developmental Stages
ESCs (n = 3) and patient-derived normal/WT (n = 3) or PiZZ (n = 3) iPSCs were analyzed at T0, T5, and T24 for a total of 27 samples.
(A) PCA demonstrates clustering predominantly by developmental stage (arrows indicate hepatic-stage PiZZ cells).
(B) Unsupervised hierarchical clustering of 27 samples analyzed by two-way ANOVA for time and cell type effects demonstrates differential gene expression with directed differentiation. The top ten genes upregulated with differentiation (middle) as well as expression levels of select endoderm genes at T5 (bottom) are listed.
(C) Analysis of gene expression differences among cell types that are modulated by differentiation (interaction effect of time and cell type) reveals 419 differentially expressed genes (FDR < 0.25; 85 genes at FDR < 0.1).
(D) Expression levels of the top five genes differentially expressed between PiZZ and normal iPSC-hepatic cells as identified by a post hoc moderated t test of the interaction effect gene set.
(E) qPCR validation of relative mRNA expression levels of genes from (D). n = 3 independent experiments. ∗p < 0.05 by two-tailed t test.
(F) qPCR quantification of ATF4 and spliced XBP-1 mRNA expression at early hepatic stage (T18) in ESCs and PiZZ or normal iPSCs. n = 3 independent experiments. ∗p < 0.05 by one-way ANOVA.
(G) Densitometric quantification of sXBP-1, Grp78, and IκBα protein expression levels by western blot of whole-cell lysates prepared from PiZZ and normal iPSCs (n = 3 biological replicates per group) at T16–T18 and normalized to GAPDH. Data are represented as mean ± SEM.
