Abstract
The mitogen-activated protein kinase (MAPK) family of genes aids cells in sensing both extracellular and intracellular stimuli, and emerging data indicate that MAPKs have fundamental, yet diverse, roles in the circadian biological clock. In the mammalian sup-rachiasmatic nucleus (SCN), MAPK pathways can function as inputs allowing the endogenous clock to entrain to 24 h environmental cycles. MAPKs can also interact physically and/or genetically with components of the molecular circadian oscillator, implying that MAPKs can affect the cycling of the clock. Finally, circadian rhythms in MAPK pathway activation exist in many different tissue types and in model organisms, providing a mechanism to coordinately control the expression tissue-specific target genes at the proper time of day. As such, it should probably not come as a surprise that MAPK signaling pathways and circadian clocks affect similar biological processes and defects in either pathway lead to many of the same types of human diseases, highlighting the need to better define the mechanisms that link these two fundamental pathways together.
1. CIRCADIAN CLOCKS
1.1. The beginning
Life on our planet is constantly challenged by changes in environmental conditions that stress the organism. To survive, organisms have developed mechanisms to aid in adapting to stress. In particular, signal-transduction pathways provide a biochemical means to sense and appropriately respond to different stress signals. This response will typically help the organism overcome an acute stress in the short term, in the span of minutes to hours. However, certain stressors mediated by the rotation of the earth, such as the UV light from the sun, daily increases in temperature and humidity, or even the presence of predators, impose a predictable stress on organisms. Without a means to anticipate these daily events, we, and other organisms, would be playing a constant game of catch up to mount an appropriate response. Alternatively, precious resources would be wasted if protection were provided at times of the day when it is not needed. It therefore makes sense that organisms have evolved a means to tell time, using an internal circadian clock to allow anticipation of environmental cycles. New data have revealed fascinating linkages between the circadian clock and the signal-transduction pathways that allow the organism to deal appropriately with rhythmic stress signals, and insights into these connections have potential for the development of new therapies to treat diseases common to both pathways.
1.2. Organization of the circadian clock
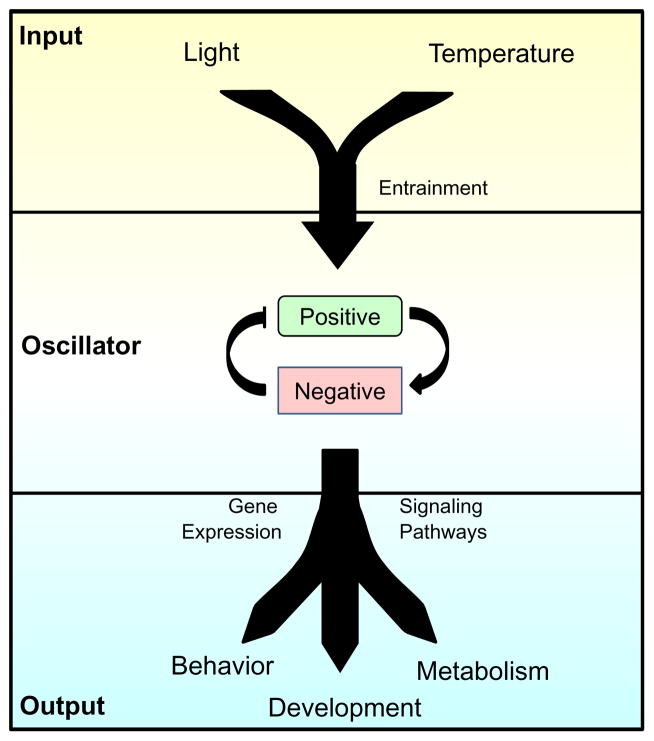
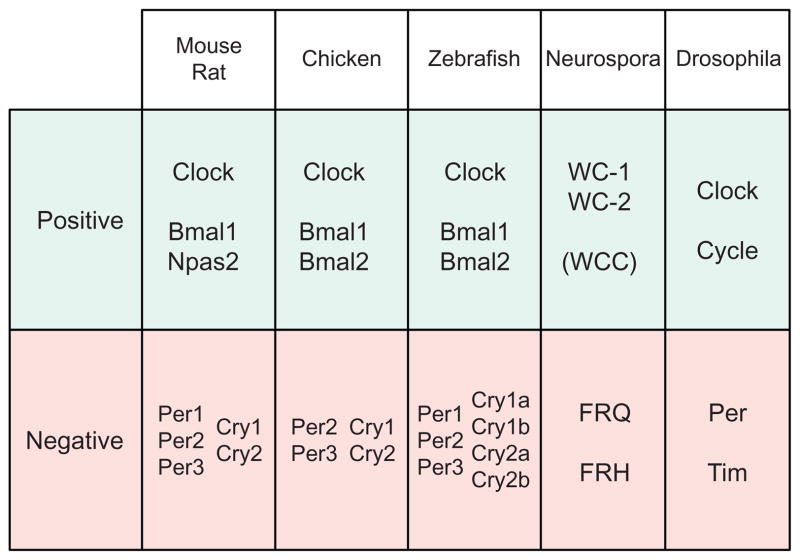
The proper functioning of the circadian clock requires the integration of many biological processes (Figure 1.1). At the core of a circadian clock, a molecular oscillator cycles with a period of around 24 h and acts as a pacemaker to generate endogenous rhythms to match the daily solar cycle. The genes that comprise the oscillators are generally not conserved across phyla (Tauber, Last, Olive, & Kyriacou, 2004). However, oscillator components are related by descent in mammals, and evolutionary relationships between the positive elements WC-1 in Neurospora and BMAL1 in mammals have been described (Lee, Dunlap, & Loros, 2000; Tauber et al., 2004) (Figure 1.2). In any case, eukaryotic oscillators share a common organizational principle: they function as negative feedback loops (Cahill, 2002; Dunlap, Loros, Liu, & Crosthwaite, 1999; Hardin, 2005; Ko & Takahashi, 2006) (Figure 1.1). Conceptually, a positive element(s), usually a transcriptional activator, induces the expression of the negative element(s). The negative element(s), upon sufficient accumulation, represses the activity of the positive element(s). The negative element(s) endures persistent phos-phorylation, which, over time, leads to their degradation. As the repression mediated by the negative components is diminished through its degradation, the positive element(s) can once again function to activate transcription, and the cycle restarts the next day (Dunlap et al., 1999).
Figure 1.1.
Organization of the circadian clock. The core of a circadian clock is its timekeeping molecular oscillator that cycles with a period of approximately 24 h. Input pathways detect temporal cues in the environment, such as light and temperature, and synchronize the molecular oscillator to external time through a process called entrainment. Finally, output pathways couple the molecular oscillator to the control of gene expression and signaling pathways. Rhythmic control of output pathways by the clock underlies overt circadian rhythms in behavior, metabolism, and development.
Figure 1.2.
Molecular components of the circadian oscillator in diverse eukaryotic organisms. Components of the molecular oscillator are categorized by their involvement in the positive or negative arm of the oscillator. Clock genes in animals show strong sequence conservation, and evolutionary relatedness between the positive elements WC-1 in Neurospora and BMAL1 in mammals has been described (Lee et al., 2000; Tauber et al., 2004).
The internal clock would be useless if it was not in synchrony with the environmental time or if the cells within a tissue were not synchronized to each other. Therefore, input pathways to the circadian oscillator are vital to maintain the proper timing of the oscillator in relation to the environment. In a process called entrainment, input pathways reset the oscillator so that the period of the oscillator conforms to the 24 h period of the environment ( Johnson, Elliott, & Foster, 2003) (Figure 1.1). Input pathways detect environmental cues and utilize various mechanisms to increase or decrease the levels or activity of a molecular oscillator component in order to set the clock to the correct time of the day. One of the most ubiquitous time-giving cues is light, but nonphotic environmental cues including temperature, nutrition, and social interactions can also entrain the circadian clock (Dunlap, 1999; Johnson et al., 2003; Mohawk, Green, & Takahashi, 2012; Mrosovsky, 1996). In addition, the clock utilizes a strategy, called gating, to restrict the response to environmental cues at certain times of the day. For example, diurnal mammals are typically insensitive to a light pulse during the day, but during the night, a light pulse can advance or delay the clock to synchronize it to the environment (Daan & Pittendrigh, 1976), just as we would adjust our watches to match the local time. In organisms of different complexity, cells vary in their ability to support a molecular oscillator that can be entrained by environmental signals. In unicellular organisms, each cell has a fully entrainable oscillator that predominantly responds to light (Bell-Pedersen et al., 2005). However, in complex multicellular organisms, not all cell types have the necessary sensory capabilities, such as photo-perception, to entrain their circadian oscillator (Mohawk et al., 2012). The cellular oscillators and overall rhythmicity of the organism are broken down into the components of the master pacemaker and peripheral oscillators (Mohawk et al., 2012). Organisms possessing a nervous system typically delegate the ability to sense environmental cues to the nervous system rather than to individual cells in order to integrate multiple sensory inputs (Mohawk et al., 2012). In general, sensory inputs to the clock are integrated in the brain, where the master pacemaker signals the entrainment of oscillators in other tissues in the organism (Mohawk et al., 2012). However, the relationship between the master pacemaker and peripheral oscillators varies between species. In Drosophila, the neuronal pacemaker is responsible for rhythms in locomotion and eclosion; however, oscillators in peripheral tissues can independently entrain to light–dark (LD) cycles (Plautz, Kaneko, Hall, & Kay, 1997). In mammals, light is perceived by nonvisual retinal ganglion cells that transmit information via neural connections to the master pacemaker, which is located in a region of the hypothalamus called the suprachiasmatic nucleus (SCN) (Dibner, Schibler, & Albrecht, 2010). The SCN pacemaker synchronizes oscillators in other tissues by a mechanism that utilizes poorly understood circadian input pathways from the SCN to individual cells in the periphery (Dibner et al., 2010). In addition to maintaining entrainment of peripheral oscillators with the environment, this system ensures that cellular oscillations within tissues are properly in phase so as to provide resonance between individual cellular rhythms. In summary, functional oscillators in individual cells require the intrinsic ability to be reset by circadian input pathways, and the master pacemaker facilitates the entrainment process in peripheral oscillators to maintain timing between cells in a tissue and between peripheral oscillators in an organism.
Another component of the circadian clock is the output pathways that connect the timekeeping oscillator, through the control of gene expression, to overt biological rhythms (Figure 1.1). In cyanobacteria, nearly the entire genome is under the control of the circadian clock through rhythmic regulation of chromosome compaction (Golden, Ishiura, Johnson, & Kondo, 1997). In the fungus Neurospora, the expression of around 20% of the genome is under the control of the clock at the level of transcript abundance (Vitalini, de Paula, Park, & Bell-Pedersen, 2006). In mouse peripheral tissues, around 10% of the transcriptome is clock-regulated, but the identity of the rhythmic transcripts varies significantly between tissue types (Storch et al., 2002). The oscillator can control the rhythmic output using different mechanisms, including direct control of gene expression by oscillator components (Koike et al., 2012; Smith et al., 2010). Many direct targets of the positive oscillator components are transcriptional regulators or signaling components, which establish a network of clock-controlled genes (CCGs) that extends beyond the direct clock targets (Smith et al., 2010) (Figure 1.1). An understanding of the circadian output has lagged compared to other areas of biological timing, partly because of the complex nature of output pathways. With new technological advances, it is now fairly straightforward to identify either direct targets of oscillator components or mRNAs that cycle in abundance. However, the ability to methodically dissect a complete regulatory network seems to be shrouded by the redundancy and complexity of cellular signaling networks. Furthermore, designating a gene as a clock output is complicated by the fact that some output genes, for example, vivid in Neurospora (Chen, DeMay, Gladfelter, Dunlap, & Loros, 2010; Smith et al., 2010) or NAMPT and SIRT1 in the mouse (Ramsey et al., 2009), feedback onto the oscillator, and therefore, they can act as both an output from and an input to the circadian clock.
2. MAPK SIGNALING PATHWAYS
2.1. General organization
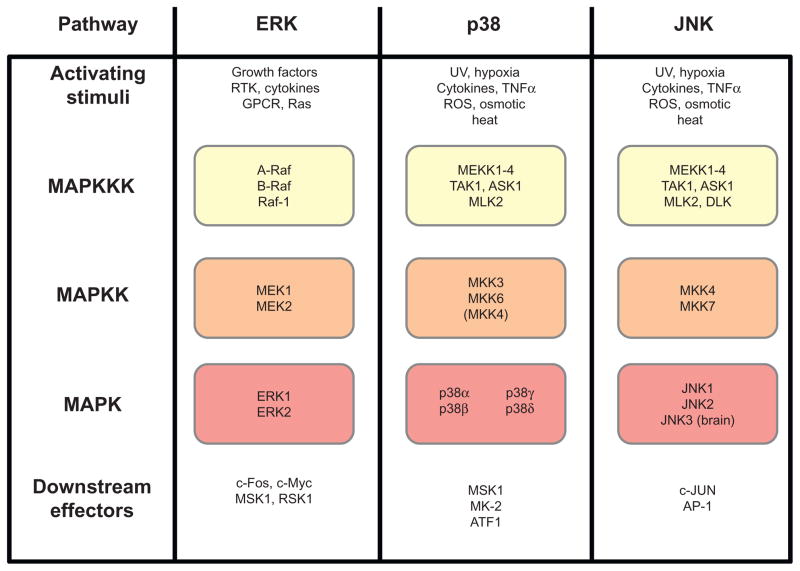
As one of the most highly conserved signaling pathways in eukaryotes, the mitogen-activated protein kinase (MAPK) pathway functions as both an output and input pathway in the circadian clock (Bennett, Beremand, Thomas, & Bell-Pedersen, 2013; de Paula, Lamb, Bennett, & Bell-Pedersen, 2008; Dziema et al., 2003; Wang & Sehgal, 2002). MAPK pathways relay intracellular and extracellular signals to mediate an appropriate response to a given mitogen (Roux & Blenis, 2004) (Figure 1.3). The MAPK signaling pathways function as kinase cascades that are composed of a canonical three-tier hierarchy of serine/threonine kinases. At the very top tier, the MAPK kinase kinase (MAPKKK) is activated by cellular signaling components in response to a stimulus. Once active, the MAPKKK phos-phorylates an associated MAPK kinase (MAPKK). Finally, the activated phospho-MAPKK couples with its cognate MAPK, the terminal component of the cascade. Once the MAPK is phosphorylated, it can associate with a host of cellular factors to control gene expression and various other cellular processes. Typically, a MAPK pathway regulates the expression of 200–500 genes depending on the activating stimulus and tissue type (Bennett et al., 2013; Gazel, Banno, Walsh, & Blumenberg, 2006; Gazel, Nijhawan, Walsh, & Blumenberg, 2008; Hagiwara et al., 2009; Roberts et al., 2000). Thus, as outputs of the circadian clock, MAPK pathways can potentially control rhythms in a large number of genes that are functionally related. Multiple MAPK pathways belonging to distinct subfamilies exist in parallel and are present in nearly all eukaryotic organisms (Johnson & Lapadat, 2002). While it simplifies matters to think of each MAPK pathway as being insulated and linear, evidence reflects a much different reality. Many of the components of signaling pathways are shared between families, and this relationship seems to increase the more upstream in the cascade that a particular component lies (Johnson & Lapadat, 2002). How a vast network of interconnected components relays and integrates information with specificity is not fully understood; however, some mechanisms are known, which allow MAPK pathways to achieve specificity. For example, scaffold proteins can physically colocalize proteins in a cascade, while excluding components of parallel MAPK pathways, in order to mediate a specific response to a stimulus (Morrison & Davis, 2003; Whitmarsh & Davis, 1998) Also, feedback inhibition, in which the activity of an MAPK leads to the inhibition of its own components or the components of a neighboring pathway, is a mechanism that contributes to signaling fidelity (Kolch, 2005; O’Rourke & Herskowitz, 1998). Often times, this inhibition is achieved through regulation of phosphatases that dephosphorylate and, therefore, inactivate MAPKs (Owens & Keyse, 2007). Three families of MAPK pathways have emerged: extracellular-signal-regulated kinase (ERK), c-Jun NH2-terminal kinase ( JNK), and p38 ( Johnson & Lapadat, 2002) (Figure 1.3). Each of these MAPK pathways has been linked directly to the circadian clock mechanism.
Figure 1.3.
Organization of MAPK pathways in mammals. Each MAPK family is organized to show the canonical signaling cascade in mammals. Activating stimuli induce the activity of the MAPKKK, which propagates a signal through the MAPKK and, finally, to the MAPK. Downstream effectors further propagate the signal to relevant molecules.
2.2. ERK MAPK
The ERK MAPKs are typified by ERK1 and ERK2 MAPKs in mammals (Roux & Blenis, 2004) (Figure 1.3). In general, ERK kinases signal in response to growth factors or, to a lesser degree, stress signals. Receptor tyro-sine kinases and G protein-coupled receptors at the cell surface activate the Ras complex, the most potent activator of ERK1/2 MAPK cascade. Ras activates Raf proteins that act as MAPKKKs in the ERK pathway (Roux & Blenis, 2004). These MAPKKKs signal to the MAPKKs, MEK1 and MEK2, which then activate ERK1/2 (Figure 1.3). ERK1 and ERK2 show 83% amino acid identity and are frequently coactivated. The activation of the ERK MAPKs is facilitated by dual phosphorylation of a Thr-Glu-Tyr motif. Downstream targets of ERK1/2 include cancer-related substrates, such as c-Fos and c-Myc, and a host of interacting regulatory kinases, including MSK-1 and RSK-1 (Figure 1.3). Thus, it is not surprising that ERKs play a key role in the regulation of cell growth and proliferation as its major upstream activator, Ras, is frequently aberrantly activated in cancer (Davies et al., 2002).
2.3. p38 MAPK
Similar to the ERK MAPK, p38 MAPK is activated by dual phosphorylation of conserved Tyr-Gly-Thr residues (Zarubin & Han, 2005). In mammals, there are four known isoforms of p38 (α, β, γ, and δ) that show tissue-specific expression; however, p38α is the most ubiquitously expressed isoform (Zarubin & Han, 2005) (Figure 1.3). p38 MAPK is tightly regulated by the MAPKK genes MKK3 and MKK6, although some signaling through MKK4, the JNK-related MAPKK, occurs (Zarubin & Han, 2005). A large network of MAPKKKs feed into the p38 MAPKs, including MEKK1–4, TAK1, ASK1, and MLK2 (Roux & Blenis, 2004) (Figure 1.3). The most classic activating signal of the p38 pathway is through the inflammatory cytokine tumor necrosis factor-α (TNF-α) and lipopolysaccharide (LPS) (Roux & Blenis, 2004), which leads to a p38-mediated inflammatory response, especially through cytokines secreted from immune cells (Zarubin & Han, 2005). Additionally, p38 in mammals and lower eukary-otes is activated by many different stress signals, including reactive oxygen species (ROS), osmotic stress, and heat shock (Zarubin & Han, 2005). DNA damage also activates p38 MAPK, which in turn regulates many genes related to cell cycle arrest and apoptosis (Zarubin & Han, 2005). Based on differential expression of p38 isoforms in different cell types, the p38 MAPK pathway provides a mechanism to address cell-type-specific needs (Zarubin & Han, 2005).
2.4. JNK MAPK
The JNK MAPK is activated by phosphorylation of the canonical MAPK activation motif Thr-Pro-Tyr (Barr & Bogoyevitch, 2001; Roux & Blenis, 2004). Of the three JNK MAPK genes, JNK1 and JNK2 are expressed in most cells, whereas JNK3 is expressed mainly in the brain (Roux & Blenis, 2004) (Figure 1.3). JNK MAPKs do not have redundant signaling roles and sometimes oppose each other’s activity (Bogoyevitch, 2006). JNK MAPKs are activated by two upstream MAPKKs, MKK4 and MKK7 (Barr & Bogoyevitch, 2001), and the top tier of the JNK cascade shares many MAPKKKs with p38 (Roux & Blenis, 2004) (Figure 1.3). Given the overlap in the upstream MAPKKK network with p38, it is not surprising that JNK is activated by many of the same stimuli as p38, with a particular sensitivity to stress signals and cytokines (Barr & Bogoyevitch, 2001). To maintain specificity of downstream signaling events, the scaffold JNK-interacting protein 1 ( JIP-1) physically associates the components of the JNK pathway (Barr & Bogoyevitch, 2001; Whitmarsh & Davis, 1998). The most recognized downstream target of the JNK MAPKs is Jun proteins involved in the activating protein 1 (AP-1) transcriptional activating complex (Hess, Angel, & Schorpp-Kistner, 2004). In contrast to the other MAPK subfamilies, JNK MAPK is not known to signal to any downstream intermediate kinases (Roux & Blenis, 2004).
3. THE FUNCTION OF MAPKs IN CIRCADIAN INPUT PATHWAYS
3.1. ERK MAPK pathway
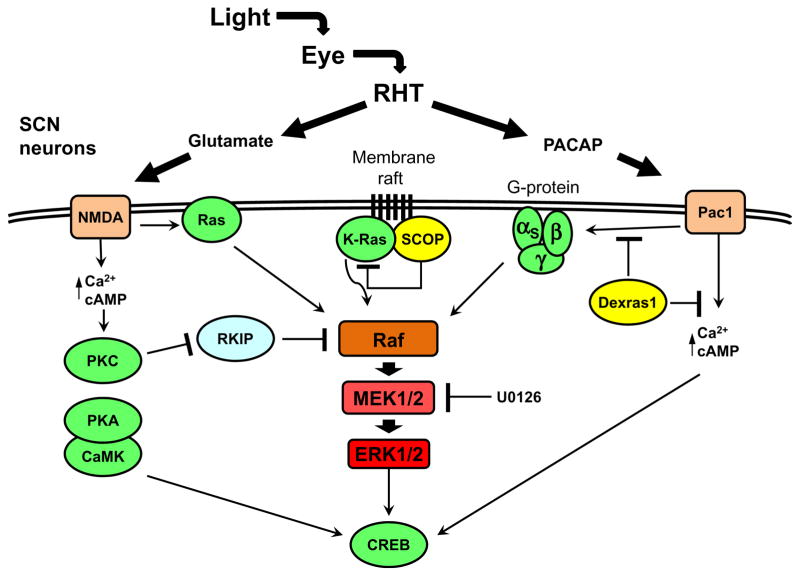
The functions of ERKs in the circadian clock are most thoroughly investigated in neural tissue, where ERKs were found to play a role in photic resetting of the clock in the rodent SCN (Butcher et al., 2002; Coogan & Piggins, 2004; Obrietan, Impey, & Storm, 1998). In order for the SCN clock to fulfill its role as a master pacemaker, the clock is entrained by light via direct innervation from the eyes. Photosensitive retinal ganglion cells (pRGC) in the eye detect light through the blue-light photoreceptor melanopsin (Schmidt, Chen, & Hattar, 2011). These neurons project directly to the SCN through the retinohypothalamic tract (RHT) where they release glutamate and pituitary adenylate cyclase-activating peptide (PACAP) that are ligands for NMDA and Pac1 receptors, respectively, at postsynaptic SCN neurons (Dibner et al., 2010) (Figure 1.4). These neurotransmitters facilitate an increase in intracellular calcium and adenosine 3′,5′-monophosphate (cAMP) that activate Ras/ERK (Dziema & Obrietan, 2002; Schurov, Hepworth, & Hastings, 2002). In addition to the activation of ERK, increases in intracellular calcium and cAMP culminate in the activation of CRE-binding protein (CREB) (Lonze & Ginty, 2002). The activation of CREB is mediated through phosphorylation at serine-133, which is a target of a myriad of kinases, including protein kinase A (PKA)/cAMP, calmodulin-dependent kinase (CaMK), and kinases downstream of the ERK MAPK (Dziema & Obrietan, 2002; Lonze & Ginty, 2002; Obrietan, Impey, Smith, Athos, & Storm, 1999; Schurov et al., 2002) (Figure 1.4). The activation of CREB in response to light, which is partly dependent on the activity of phosphorylated-ERK (p-ERK), stimulates transcriptional activation through CRE cis-elements on the promoters of target genes (Lonze & Ginty, 2002; Obrietan et al., 1998) (Figure 1.4). The immediate early genes (IEGs) are targets of CREB, and they are upregulated following photic stimulation in the SCN (Ginty et al., 1993). Many of the IEGs, notably c-Fos and Per1, require a functional ERK pathway for their light-induced expression (Dziema et al., 2003). Per1 is a clock gene in the negative arm of the circadian oscillator, and its induction is thought to be a primary event in the resetting of the clock (Tischkau, Mitchell, Tyan, Buchanan, & Gillette, 2003). Consistent with a role for ERK MAPK in light signaling to the clock, infusion of ERK inhibitor into the mouse SCN prevented phase shifts in activity rhythms when given during the subjective night (Butcher et al., 2002).
Figure 1.4.
Diagram of the light-responsive ERK signaling network in the SCN. In light-mediated clock resetting, the neurotransmitters glutamate and PACAP are released onto SCN neurons via the eye and RHT. The NMDA and Pac1 receptors lead to the activation of ERK through Ras protein and heterotrimeric G proteins, and proteins like RKIP, SCOP, and Dexras1, as well as the pharmacological inhibitor U0126, modulate the activation of ERK. The activation of ERK in response to light, along with parallel signaling pathways, leads to the activation of CREB, which is responsible for the induction of the immediate early genes, including mPer1. Components that are rhythmically expressed are shown in yellow.
An important characteristic of ERK as an input to the clock is that its response to light is gated to the subjective night (Obrietan et al., 1998). This gating is mediated through Dexras1, a Ras-like G protein expressed in the SCN during the subjective night (Cheng et al., 2004). Dexras1 regulates the light sensitivity of the clock at different times of the day by repressing or activating different signaling pathways, including repressing the ability of the neuropeptide PACAP to control ERK activity in the SCN during the late night (Cheng et al., 2004, 2006) (Figure 1.4).
3.2. ERK signaling through downstream components
ERK MAPK has an array of downstream effector molecules, such as transcription factors, kinases, and translational regulators, that control the expression of target genes, including clock genes (Roux & Blenis, 2004) (Figure 1.5). The transcription factor Elk-1 is downstream of ERK, and its activity, regulated by ERK-dependent phosphorylation, leads to transcriptional activation of target genes (Davis, Vanhoutte, Pagès, Caboche, & Laroche, 2000). Elk-1 is phosphorylated in response to a light pulse given at night, and this is dependent on a functional ERK pathway (Coogan & Piggins, 2003). Phosphorylated Elk-1 (p-Elk-1) is also induced by glutamate treatment, a crucial neurotransmitter in clock resetting. Following activation, p-Elk-1 binds the serum response element (SRE) in the promoter of target genes that include c-Fos and Per1 (Davis et al., 2000; Vanhoutte et al., 1999) (Figure 1.5). Furthermore, bioinformatic analysis in mouse liver has shown that the SRE is enriched in the promoters of CCGs (Bozek, Rosahl, Gaub, Lorenzen, & Herzel, 2010). Thus, given the established role of Elk-1 as a transcription factor downstream of ERK, it is likely that Elk-1 plays an important role in regulating ERK target genes in the SCN in response to light, including core oscillator genes.
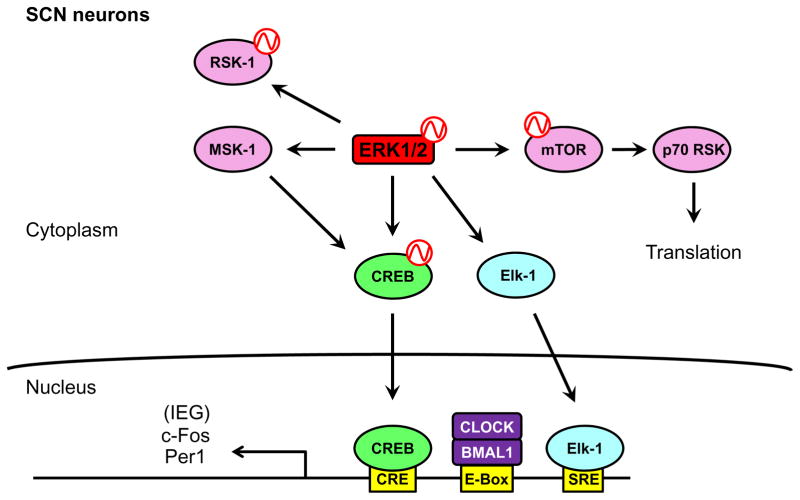
Figure 1.5.
Downstream components of ERK in SCN neurons. Activated ERK can signal through various downstream components in the SCN. The intermediate kinases RSK-1 and MSK-1 are activated by ERK after light stimulus. Also, ERK signals through the mTOR/ p70 RSK pathway to putatively regulate translation of target genes. Finally, ERK activates the CREB and Elk-1 transcription factors to induce the expression of the immediate early genes in response to light stimulus. Rhythmicity in the activity of a protein is indicated by a red sine wave.
In addition to Elk-1, ERK interacts with a network of downstream effector kinases (Anjum & Blenis, 2008; Roux & Blenis, 2004) (Figure 1.5). For example, the p90 ribosomal S6 kinase 1 (RSK-1) is an ERK-regulated kinase that requires a functional ERK pathway for light-induced phosphorylation in the SCN, and RSK-1 directly phosphorylates CREB (Butcher, Lee, Hsieh, & Obrietan, 2004; Lonze & Ginty, 2002; Xing, Ginty, & Greenberg, 1996) Another intermediate effector kinase, mitogen- and stress-activated protein kinase 1 (MSK-1), is thought to act as an intermediate between ERK and p-CREB. In the mouse SCN, active phosphorylated MSK-1 colocalizes with ERK-active neurons following a light pulse or after administration of PACAP (Butcher, Lee, Cheng, & Obrietan, 2005). In support of the hypothesis that MSK-1 functions in the ERK circadian input pathway, light pulses given to an MSK-1 knockout mouse showed diminished phase shifts and lower levels of the necessary gene products for clock resetting, including phosphorylated CREB (p-CREB), c-Fos, and Per1 (Cao, Butcher, Karelina, Arthur, & Obrietan, 2013).
The ERK MAPK also couples with downstream processes that are unrelated to transcriptional activation. Through micro-RNA (miRNA) expression, the ERK/CREB signaling module can regulate gene expression at the posttranscriptional level. miR-132 is a light-induced miRNA in the mouse SCN that requires a functional ERK pathway and CREB signaling for its expression (Cheng et al., 2007). The blockage of miR-132 activity leads to reduced phase shifting after a light pulse, and miR-132 enhances Per1 protein expression after a light pulse (Cheng et al., 2007). In addition, the mammalian target of rapamycin (mTOR) pathway, a master regulator of translation, plays a significant role in the SCN clock (Cao, Anderson, Jung, Dziema, & Obrietan, 2011; Cao, Lee, Cho, Saklayen, & Obrietan, 2008; Cao, Li, Cho, Lee, & Obrietan, 2010) (Figure 1.5). The activity of mTOR is rhythmic in the SCN (Cao et al., 2011), and inhibition of mTOR activity leads to defective light-induced phase shifting of mouse activity rhythms (Cao et al., 2010). The mTOR-regulated kinase p70 RSK is activated in ERK-active neurons in the SCN after a light pulse, and the inhibition of either ERK or mTOR suppresses light-induced p70 RSK activation (Cao et al., 2008, 2010). These data suggested that an ERK/mTOR/p70 RSK signaling pathway regulates translation in response to photic stimulation in the SCN (Figure 1.5). Because p70 RSK colocalizes with p-ERK and p-CREB following a light pulse, it was suggested that upon a light pulse at night, ERK coordinates transcriptional and translational activation to induce the expression of light-responsive genes, providing a mechanism to synchronize the clock in the SCN to the environment (Cao et al., 2008).
3.3. ERK MAPK in peripheral tissues
A role for ERK MAPK in the light input pathway to the circadian oscillator is most clearly defined in the SCN. Limited data exist for ERK having a role in circadian input pathways of peripheral tissues in mammals (Akashi & Nishida, 2000); however, insights from model organisms support a role for ERK in resetting in peripheral oscillators. For example, the zebra fish Z3 peripheral cell line retains the ability to detect light, and the photic stimulation of Z3 cells induces the expression of the clock gene zPer2 via ERK activation (Cermakian et al., 2002). Consistent with the role for ERK in resetting of mammalian peripheral clocks, stimuli known to reset the oscillator in in vitro mammalian cultures, such as treatment with dexamethasone, forskolin, or concentrated serum, induce signaling pathways common to photic stimulus in the SCN and lead to the activation of the same immediate early genes (Balsalobre, Marcacci, & Schibler, 2000). Indeed, after certain stimuli, ERK activity is necessary for the induction of clock resetting genes in NIH-3T3 fibroblasts, but ERK does not affect the oscillation of circadian clock gene expression (Akashi & Nishida, 2000). While these data support a role for ERK in the circadian input pathway of peripheral oscillators, further investigation is needed.
3.4. p38 MAPK pathway
In contrast to ERK MAPK, p38’s role in photic clock resetting is less clear and appears to be dispensable. In rodents, p38 MAPK is activated in response to light in the SCN during the subjective night, matching the phase gating of ERK in the hamster SCN (Pizzio, Hainich, Ferreyra, Coso, & Golombek, 2003). In cultured rat pineal gland, p38 MAPK activation is similar to ERK in that both are rhythmic in an LD cycle, peaking during the subjective night (Chik, Mackova, Price, & Ho, 2004; Ho, Mackova, Price, & Chik, 2003). However, blockage of p38 signaling had no effect on the activation of CREB in cortical neurons, demonstrating that p38 probably does not signal through the CREB-mediated clock resetting pathway (Arthur et al., 2004).
In the chick pineal gland, p38 MAPK may play a role in photic resetting of the clock-controlled rhythm in melatonin production; brief treatment with p38 inhibitor SB 203580 generated dark pulse-like phase shifts (Hayashi, Sanada, Hirota, Shimizu, & Fukada, 2003; Yadav, Straume, Heath, & Zatz, 2003) (Figure 1.6). Similarly, in cultured Xenopus retina, p38 MAPK appeared to play a role in photic resetting of the clock-controlled rhythm in melatonin production (Hasegawa & Cahill, 2004). Administration of p38 inhibitor SB 203580 had no acute effect on the production of melatonin, but brief treatments of cultured retinas with SB 203580 led to phase shifts in the melatonin rhythm (Hasegawa & Cahill, 2004) (Figure 1.6). Together, these data suggested that p38 does not signal in an output pathway controlling melatonin rhythms, but, instead, is part of an input pathway that modulates the endogenous oscillator. However, SB 203580 has nonspecific interactions with other kinases, including JNK MAPK and CKIε, which may account for the inhibitor-induced phase shifts (Hasegawa & Cahill, 2004), particularly because CK1ε is known to modulate the activity of components of the circadian oscillator (Kloss et al., 1998; Lowrey et al., 2000; Price et al., 1998). Furthermore, JNK inhibition with the drug SP600125 in zebra fish leads to dark pulse-like phase shifts of the melatonin rhythm that are similar to those elicited by CKIε-specific inhibition (Hasegawa & Cahill, 2004). Thus, while the inhibitor studies suggested a conserved role for the p38 MAPK pathway in circadian input in neural tissues, additional studies are needed using specific p38 inhibitors, such as VX-745 (Bagley, Davis, Dix, Rokicki, & Kipling, 2007; Fabian et al., 2005), or through genetic disruption.
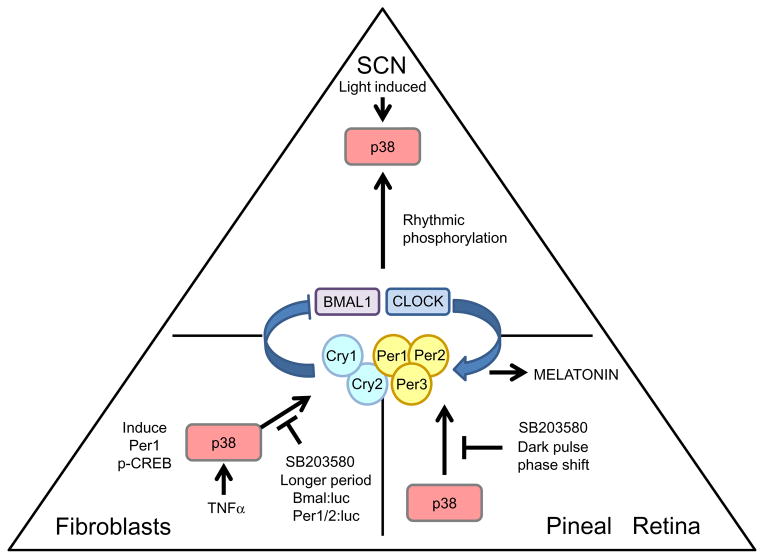
Figure 1.6.
The role of p38 MAPK in the circadian clock. The diagram presents the most representative tissues in mammals with regard to the function of p38—either SCN, fibroblasts, or the pineal/retina. The oscillator in the center of the triangle represents the cellular oscillator in the respective tissues. The arrows that are drawn toward the oscillator represent a circadian input pathway. The lines that are drawn away from either the oscillator or p38 MAPK represent output pathways. The labels indicate the functions of pathways or perturbations on the activity of p38.
3.5. p38 MAPK in peripheral tissues
The mammalian p38 MAPK pathway is thought to impinge upon the circadian oscillator after activation by the inflammatory cytokine TNF-α (Petrzilka, Taraborrelli, Cavadini, Fontana, & Birchler, 2009; Zarubin & Han, 2005) (Figure 1.6). TNF-α in humans has been implicated in daytime fatigue (Spriggs et al., 1988), and accordingly, inflammatory disease-associated fatigue seems to be suppressed by treatment with TNF-α antagonists (Pollard, Choy, Gonzalez, Khoshaba, & Scott, 2006; Vgontzas et al., 2004). To investigate a putative role for TNF-α in circadian disruption, mouse fibroblasts were used to measure the circadian response to TNF-α. The induced expression of Per1 through CREB-mediated transcription is a critical event in clock resetting, and the treatment of fibroblasts with p38 inhibitor SB 203580 effectively blocked TNF-α-mediated induction of both p-CREB and Per1 (Petrzilka et al., 2009). While TNF-α induced Per1 expression via p38 activation, TNF-α treatment of fibroblasts was not sufficient to induce persistent circadian rhythms (Petrzilka et al., 2009). These data suggested that while p38 may be involved in the induction of clock genes by TNF-α, it is not sufficient to reset the clock. Clearly, the use the nonspecific inhibitor SB 203580 needs to be taken into account when interpreting the results of this study; however, these data raise the intriguing possibility that p38 modulates circadian rhythms in immune cells.
p38 MAPK has also been implicated in circadian input that couples humoral signals arising from the SCN to individual oscillators in peripheral tissues (Ko et al., 2011). Wild-type adult mice maintain a rhythm in phos-phorylated p38 (p-p38) within cardiac tissue when housed in an LD cycle (Ko et al., 2011). Furthermore, a cardiac-specific expression of a dominant null allele of Clock abolishes the circadian oscillator specifically in cardiomyocytes, but the peripheral oscillators outside of the heart remain intact (Bray et al., 2008). Interestingly, in mice with cardiac-specific disruption of the circadian oscillator housed in an LD cycle, diurnal rhythms of p38 activity are maintained in the heart, suggesting that p38 relays a rhythmic extracellular signal from the SCN (Ko et al., 2011). However, it remains unclear if a functional clock in the SCN is necessary for the observed rhythms in p38 in clock-disrupted cardiomyocytes or if rhythmicity results from light- or activity-driven rhythms.
3.6. JNK MAPK pathway
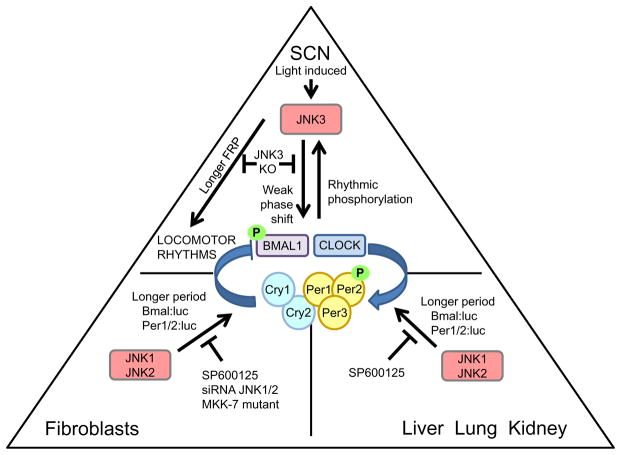
In the hamster and mouse SCN, a light pulse at night leads to phosphory-lation of JNK (Pizzio et al., 2003; Yoshitane et al., 2012). Additionally, in rat-1 fibroblasts, a media change that resets the clock also leads to phosphor-ylation of JNK MAPK (Chansard, Molyneux, Nomura, Harrington, & Fukuhara, 2007). In JNK3 knockout mice, a light pulse given during the subjective night results in a diminished phase shift, indicating a defect in circadian entrainment (Yoshitane et al., 2012) (Figure 1.7). Interestingly, the JNK3 knockout does not have any effect on the light-induced transcription of Per1 in the SCN, suggesting that the JNK MAPK does not participate clock resetting through CREB-mediated Per1 activation (Yoshitane et al., 2012). In support of this idea, activated JNK MAPK was shown to phosphorylate both CLOCK and BMAL1 (Yoshitane et al., 2012). These data imply that JNK is involved in clock resetting; however, instead of inducing Per1 expression, its mechanism relies primarily on the phosphor-ylation of clock proteins, which is discussed further in the succeeding text.
Figure 1.7.
The role of JNK MAPK in the circadian clock. The diagram presents the most representative tissues in mammals with regard to the function of JNK—the SCN, fibro-blasts, or liver, lung, and kidney. The oscillator in the center of the triangle represents the cellular oscillator in the respective tissues. The arrows that are drawn toward the oscillator represent a circadian input pathway. The lines that are drawn away from either the oscillator or the JNK MAPK represent output pathways. The labels indicate the functions of pathways or perturbations on the activity of JNK.
3.7. Summary
In mammals, each MAPK pathway subfamily serves as input pathways to the clock in distinctly different ways. The ERK MAPK pathway facilitates clock resetting in the SCN in response to light via CREB induction of clock genes, and evidence exists to support ERK functioning in input pathways to clocks in peripheral tissues. On the other hand, p38 MAPK does not facilitate clock resetting in the SCN, but appears to signal through the same CREB-mediated clock resetting pathway in peripheral tissues. Finally, the JNK pathway contributes to photic clock resetting in the SCN, but this action is facilitated through a CREB-independent mechanism.
4. ENDOGENOUS RHYTHMS IN MAPK ACTIVATION
4.1. ERK in the SCN
Light-induced activation of ERK is important for the resetting of the mammalian circadian oscillator in response to light; however, the activation of ERK also has an endogenous rhythm in the SCN (Obrietan et al., 1998; Pizzio et al., 2003). While the phosphorylation state of ERK, and thereby its activity, displays a robust circadian rhythm, ERK protein levels are constitutive, indicating that clock regulation of ERK activation is posttransla-tional (Obrietan et al., 1998). Curiously, p-ERK activation rhythms vary between different anatomical regions of the SCN (Nakaya, Sanada, & Fukada, 2003; Obrietan et al., 1998). In the dorsomedial sector of the SCN, known as the shell, ERK activation cycles and peaks during the subjective day (Nakaya et al., 2003; Obrietan et al., 1998). In contrast, in neurons located in the ventrolateral region of the SCN, known as the “core,” p-ERK activity cycles with a peak during the subjective night (Nakaya et al., 2003; Obrietan et al., 1998). The SCN core receives direct afferents from the RHT, and nighttime photic stimulation results in p-ERK induction only in the SCN core, consistent with the core receiving neuronal input directly from the retina (Lee, Nelms, Nguyen, Silver, & Lehman, 2003; Nakaya et al., 2003). In enucleated mice, p-ERK does not accumulate in the SCN core, but continues to cycle in the SCN shell (Lee et al., 2003), indicating that rhythms in p-ERK in the SCN shell are not dependent on the SCN core or on input from the retina (Lee et al., 2003). In addition to rhythms of p-ERK in the SCN, p-ERK appears to cycle in the mouse liver (Tsuchiya, Minami, Kadotani, Todo, & Nishida, 2013). Together, these data support p-ERK activity being controlled by the clock in both pacemaker and peripheral cells and back a role for p-ERK, particularly in the SCN core, for light resetting of the clock.
4.2. p38 and JNK MAPK
Both JNKs and p38 MAPKs are reported to have endogenous activity rhythms in DD in the hamster SCN, with peaks during the late subjective day, and daytime activation in an LD cycle (Pizzio et al., 2003). In addition, JNK MAPK displays an endogenous activity rhythm in synchronized rat-1 cells (Chansard et al., 2007). Our studies reveal that p38 activity rhythms exist in cultured SCN and in peripheral tissues (Goldsmith, Moon, Earnest, and Bell-Pedersen, unpublished data). Tissue-specific rhythmic activity of the MAPKs, and their downstream effectors, may provide a mechanism to control large sets of CCGs with tissue-specific functions and offers a possible rationale for the disparity observed among CCGs in different tissues (Storch et al., 2002).
4.3. Transcriptional regulation of MAPK components
The endogenous rhythms in MAPK activation occur in the absence of environmental stimuli; thus, circadian control of MAPK activation is not a direct function of a role for MAPKs in input pathways that respond to entraining signals, although rhythmic MAPK activation may affect gating of environmental cues (see the succeeding text). Several studies have indicated that the circadian oscillator regulates the transcription of MAPK pathway components and upstream regulators as a mechanism to generate rhythms in MAPK activation. For example, evening-specific transcription of SCN circadian oscillatory protein (SCOP) is thought to be necessary to generate rhythmic ERK MAPK activation in the rodent SCN (Shimizu, Okada, Nagai, & Fukada, 2003; Shimizu, Okada, Takano, & Nagai, 1999). The SCOP localizes to a membrane raft and binds K-Ras preventing GTP binding, thereby preventing Ras activation in the early subjective day and driving daytime rhythms in ERK activation (Shimizu et al., 2003) (Figure 1.4). It is not yet known if SCOP is selectively expressed in distinct anatomical regions of the SCN and thereby contributes to the phase differences of p-ERK rhythms observed in the shell and core.
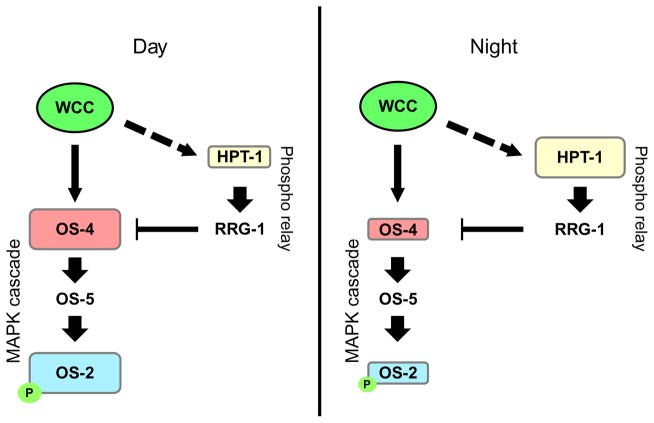
Due to the conservation of MAPK pathways across eukaryotes, model organisms provide a powerful tool to study the circadian regulation of MAPK pathways. In Neurospora, the p38 homolog, OS-2, is rhythmically phosphorylated in DD with a peak during the early subjective morning, and this rhythm requires a functional circadian oscillator (Vitalini et al., 2007). The os-4 gene, encoding the MAPKKK, is a direct target of the positive component of the Neurospora circadian oscillator, the White Collar Complex (WCC) (Figure 1.8). Rhythmic binding of the WCC to the promoter of os-4 drives oscillations in os-4 mRNA (Lamb, Goldsmith, Bennett, Finch, & Bell-Pedersen, 2011). Importantly, the circadian rhythm of phosphorylated OS-2 (p-OS-2) is a dependent rhythmic transcription of os-4, demonstrating that transcriptional activation of a MAPKKK can generate endogenous rhythms in MAPK activation (Lamb et al., 2011). Additionally, hpt-1, a gene that encodes an upstream inhibitor of the OS-2 MAPK pathway and is a component of a fungal phosphorelay that responds and signals an acute osmotic stress to the organism, is rhythmically expressed with a peak during the subjective night, the phase opposite of the os-4 mRNA and p-OS-2 rhythms (Lamb et al., 2011). Based on these findings, it was suggested that transcriptional regulation of OS MAPK pathway component genes by the circadian clock, os-4 and hpt-1, at opposite phases, modulates the abundance of the corresponding gene products, which in turn leads to rhythms in p-OS-2 (Lamb et al., 2011) (Figure 1.8). The model posits that transcriptional regulation by the circadian clock segregates the expression of positive and negative regulators into opposite phases, thereby creating resonance of their activities. This resonance is thought to tune the levels of basal signal transduction in the OS pathway and produce endogenous rhythms of p-OS-2 (Lamb et al., 2011). Rhythmic OS-2 activity leads to rhythms in downstream effector molecules that control the expression and activity of target genes in the pathway, helping the fungus to survive predictable abiotic stress signals that result from the Earth’s rotation (Lamb, Finch, & Bell-Pedersen, 2012).
Figure 1.8.
Transcriptional regulation of the OS MAPK pathway by the circadian clock in Neurospora. The genes hpt-1 and os-4 are rhythmically expressed genes, and they have opposing effects on the activation state of the OS pathway, measured by p-OS-2. The WCC, the positive oscillator component, directly regulates the expression of os-4, but the expression of hpt-1 is mediated through an indirect mechanism. It is hypothesized that the clock coordinates the antiphasic expression of these two genes to generate rhythms in p-OS-2. During the day, the WCC directly upregulates the expression of os-4, while hpt-1 is at a trough in its rhythmic expression, leading to an increase in p-OS-2. Conversely, during the night, os-4 is at a trough in its rhythmic expression, while hpt-1 is at its peak, leading to lower levels of p-OS-2.
4.4. Transcriptional regulation of MAPK components in mammalian peripheral tissue
A review of genome-wide circadian gene expression data sets in various mammalian tissues reveals mRNA rhythms in MAPK pathway components, although these have not been validated by independent means. For example, the genes Mkk3, Mkk6, and Mkk4, encoding MAPKKs upstream of p38 and JNK kinases, have a circadian rhythm in mRNA accumulation in the mouse liver (Koike et al., 2012; Menet, Rodriguez, Abruzzi, & Rosbash, 2012). Also, a variety of genes encoding MAPKs and MAPKKKs display rhythmic expression in mouse liver (Koike et al., 2012; Menet et al., 2012; Vollmers et al., 2012) and macrophages (Keller et al., 2009). These data provide promising indications that MAPK activity is rhythmically activated in a variety of mammalian tissues and that these rhythms result from clock control of transcription, similar to what has been observed in the mouse SCN and in Neurospora (Lamb et al., 2011; Shimizu et al., 1999).
4.5. Rhythmic MAPK activity gates input to the circadian clock
The biological function of circadian MAPK activity rhythms can be manifold. Rhythmic MAPK activation can serve as a circadian output pathway, controlling rhythms in target gene expression, or alternatively, it can feedback to regulate the molecular oscillator. In addition, MAPK activity rhythms can function in gating circadian input pathways. For example, in chick retina, ERK is rhythmically phosphorylated with a peak during subjective night (Ko, Ko, & Dryer, 2001). This ERK activity rhythm, along with CaMKII, modulates the membrane potential of photoreceptive cone cells through cGMP-gated cation channels, so as to regulate the photosensitivity of retinal neurons based on the time of the day (Ko et al., 2001). Intriguingly, the circadian regulation of p-ERK is dominant to photic stimulation in chick retina, with p-ERK activation being solely dependent on circadian phase (Ko, Shi, & Ko, 2009). Thus, clock control of p-ERK activation in the retina results in limiting the sensitivity of retinal photoreceptors that are vital for entrainment to specific times of the day.
5. MAPKs MODULATE THE CIRCADIAN OSCILLATOR
5.1. ERK-mediated regulation of the circadian oscillator
ERK MAPK can directly interact with components of the circadian oscillator, and ERK-mediated phosphorylation of these proteins likely plays an important role in the maintenance of biological rhythms (Akashi, Hayasaka, Yamazaki, & Node, 2008; Sanada, Harada, Sakai, Todo, & Fukada, 2004; Sanada, Okano, & Fukada, 2002; Weber, Hung, Maurer, & Kay, 2006) (Figure 1.9). Mass spectrometry identified several BMAL1 phosphorylation sites that are targets of ERK, and these phosphorylation sites facilitate the repression of CLOCK/BMAL transactivation in vitro (Sanada et al., 2002). Likewise, the Drosophila CLOCK gene, which heterodimerizes with CYCLE to form the positive component of the Drosophila circadian feedback loop (Hardin, 2005), is phosphorylated by ERK2 in cultured cells (Weber et al., 2006). The negative clock components of the mammalian clock, CRY1 and CRY2, also directly interacted with ERK in COS7 cells, and phosphorylation sites on both CRY1 and CRY2 were determined by mass spectrometry to be specific targets of ERK (Sanada et al., 2004) (Figure 1.9).
Figure 1.9.
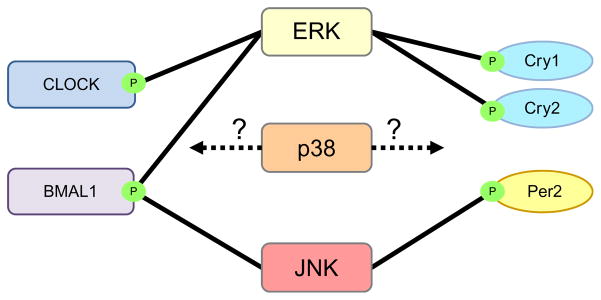
Interaction of MAPKs with clock proteins. MAPKs from different families can directly influence clock proteins in the molecular oscillator. ERK MAPK physically interacts with and phosphorylates CLOCK and BMAL1 in the positive branch of the oscillator and CRY1 and CRY2 in the negative branch. No physical interactions of p38 with clock proteins have been observed, and no p38-specific phosphorylation sites have been identified on clock proteins. However, inhibition of p38 led to a longer period of clock gene oscillation, implying that p38 may have an unknown effect on the molecular oscillator. Finally, JNK MAPK interacts with and phosphorylates BMAL1 and PER2.
Given that ERK can directly phosphorylate the gene products of the molecular oscillator, one might expect manipulation of ERK activity to cause defects in endogenous circadian rhythms. However, chronic defects in ERK signaling did not lead to defects in biological rhythms in constant conditions (Antoun, Cannon, & Cheng, 2012; Butcher et al., 2002; Hainich, Pizzio, & Golombek, 2006). In the whole animal, administration of the ERK inhibitor U0126 to a mouse or, conversely, constitutive activation of ERK in a hamster had no effect on the free-running locomotor rhythm of rodents in constant conditions (Butcher et al., 2002; Hainich et al., 2006). Similarly, in transgenic mice with a constitutively active allele of RKIP, ERK signaling is suppressed in the SCN, but no defect in the free-running locomotor rhythm was observed (Antoun et al., 2012) (Figure 1.4). In a more direct approach, cultured mouse SCN was bathed with the ERK inhibitor U0126 leading to a rapid decrease in amplitude of PER2 protein rhythms, as well as a decrease in overall Bmal1, Per1, and Per2 expression levels (Akashi et al., 2008). However, the rhythmicity of clock gene expression persisted with a wild-type phase despite the lack of ERK signaling (Akashi et al., 2008). It is possible that the disruption of ERK signaling is negligible toward oscillator function since both positive and negative components of the oscillator can be ERK targets, thereby offsetting its effect. While ERK activity can affect the amplitude of clock gene oscillations, it appears to be dispensable for the cycling of the molecular oscillator, supporting the idea that the main role for ERK in the clock system is through its activity in circadian light entrainment.
5.2. p38-mediated regulation of the circadian oscillator
Several reports suggested that the activity of p38 affects the circadian oscillators’ ability to maintain endogenous rhythms (Figure 1.6). In the chick pineal gland, chronic treatment with the SB 203580 inhibitor lengthened the endogenous period of melatonin production (Hayashi et al., 2003). A similar lengthening of the period of the clock gene rhythms was described in U2OS cells and C6 mouse glioblastoma cells after treatment with SB 203580 and SB 202190 inhibitors (Yagita, Yamanaka, Koinuma, Shigeyoshi, & Uchiyama, 2009). Similar to the ERK MAPK, p38 may phos-phorylate clock oscillator proteins, thereby affecting the stability of these proteins and altering the cycling of the molecular oscillator, although no specific interactions between p38 and clock proteins have been described (Figure 1.9). Another possibility is that the increase in period is due to CKIε off target effects as was previously discussed. Thus, these experiments need to be repeated using a specific pharmacological inhibitor or through genetic tools, such as siRNA and hypomorphic p38 alleles.
5.3. JNK-mediated regulation of the circadian oscillator
Studies show that JNK kinase acts as an input to the circadian clock independently of CREB-mediated clock resetting, and this input is likely achieved through interaction with clock proteins (Yoshitane et al., 2012) (Figure 1.7). JNK kinase modulates the properties of the circadian oscillator by phosphorylating the clock proteins in both the positive and negative branches of the oscillator (Uchida et al., 2012; Yoshitane et al., 2012) (Figure 1.9). BMAL1 protein, which forms the positive component of the mouse oscillator upon heterodimerization with CLOCK, interacts with JNK in vitro (Uchida et al., 2012). The phosphorylation of BMAL1 increases following activation of the JNK pathway and decreases following inhibition of JNK activity in in vitro cell culture (Uchida et al., 2012; Yoshitane et al., 2012). In the negative branch of the circadian oscillator, JNK MAPK interacts with PER2, and its activity increases PER2 phosphorylation (Uchida et al., 2012) (Figure 1.9). Importantly, the phosphorylation of PER2 by JNK has a significant effect on PER2 protein levels and stability (Uchida et al., 2012). Consistent with these data, an expression of a hyperactive JNK MAPK in mouse embryonic fibroblasts (MEFs) decreases the ubiquitination of PER2, indicating that JNK activity increases PER2 protein stability by decreasing its degradation via the proteasome (Uchida et al., 2012). These data demonstrate that JNK-mediated modulation of the circadian oscillator relies on the phosphorylation of clock proteins, which in turn affects the clock protein stability.
To understand the effect of JNK MAPK on the function of the molecular oscillator, initial studies focused on testing the effects of JNK inhibition on the clock. The JNK inhibitor SP600125 lengthens the period of melatonin production in chick pineal gland and bullfrog retina (Bennett et al., 2001; Hasegawa & Cahill, 2004; Hayashi et al., 2003) (Figure 1.7). Also, in rat-1, NIH-3T3, and C6 mouse glioblastoma cell lines, the treatment with JNK inhibitor results in a long period (>8 h over untreated) of clock gene oscillations (Chansard et al., 2007; Yagita et al., 2009; Yoshitane et al., 2012). Furthermore, mouse SCN, pineal, and lung explant cultures had similarly long periods of clock gene oscillations after SP600125 treatment (Chansard et al., 2007) (Figure 1.7). While the long period observed in these studies are consistent with a role for JNK in PER2 protein stability (i.e., increased stability of a negative component of the oscillator would result in a long period), similar to the p38 inhibitors, SP600125 affects a broad range of other kinases, including CKI (Bain, McLauchlan, Elliott, & Cohen, 2003; Fabian et al., 2005). Fortunately, several genetic disruptions have validated the effect of pharmacological inhibition of JNK activity on clock gene oscillations, although the effect was much less severe. Knockdown of JNK1/2 expression by siRNA in NIH-3T3 cells and a conditional knockout of MKK7, the upstream MAPKK, in MEFs led to a 1–2 h longer period of clock gene oscillations compared to wild-type cells (Uchida et al., 2012; Yoshitane et al., 2012) (Figure 1.7). These data suggest that, along with its role in resetting of the phase of the clock in response to light, JNK activity modulates the period of the oscillator by affecting the stability of PER2 and possibly by altering the activity or levels of Bmal1.
5.4. Future directions
The phosphorylation of clock proteins by MAPKs and the resulting biochemical effects by MAPKs need further investigation. Interestingly, clock protein interactions with p38 have not been observed, despite the evidence that inhibition of p38 leads to a long period of clock gene oscillations, suggesting that the effects are indirect. However, the long period caused by inhibition of p38 needs to be validated with more specific inhibitors and siRNA knockdown. On the other hand, ERK-specific phosphorylation sites have been identified on clock proteins, but surprisingly, inhibition of ERK activity does not appear to affect the endogenous rhythms of the molecular oscillator. A better understanding of ERK-mediated phosphory-lation of clock proteins in vivo may provide better insight into this observation. The effect of JNK phosphorylation of clock proteins is the most thoroughly understood, although the specific effect of JNK activity on Bmal1 requires further investigation.
One key question regarding MAPK interaction with the molecular oscillator is if MAPK phosphorylation of clock proteins only occurs following an acute environmental stimulus and/or if MAPKs phosphorylate oscillator components rhythmically in constant conditions as a function of their endogenous circadian rhythm in activation. The latter scenario implies that MAPK pathways feedback onto the oscillator as an accessory feedback loop to add robustness to the oscillator. The ability of MAPK pathways to interact with oscillator components in both peripheral and master pacemaker tissues validates the need to further study the role of MAPKs in the function of circadian oscillators in constant conditions.
6. THE ROLE OF MAPK PATHWAYS IN CIRCADIAN OUTPUT
Although components of output pathways frequently exert some feedback onto the oscillator (Roenneberg & Merrow, 1998), in general, output pathways relay temporal information from the timekeeping oscillator to terminal target genes. This definition assumes that disruption of an output pathway will disrupt rhythmicity of CCGs without affecting the function of the oscillator. If MAPK activity rhythms are indeed maintained in peripheral tissues, it would seem logical to assume that the MAPK pathway is an output pathway of the clock in these cells.
6.1. ERK MAPK pathway
The pineal gland, a neuroendocrine component of the clock that contributes to overall rhythmicity of the organism, produces the hormone melatonin that is secreted at night into the circulatory system to relay temporal information to peripheral oscillators (Dibner et al., 2010; Ho et al., 2003). In the rat pineal gland, the ERK pathway is activated by the sympathetic nervous system and is rhythmically activated in an LD cycle with a peak during the subjective night (Ho et al., 2003). In this tissue, ERK regulates the synthesis of a rate-limiting precursor in melatonin synthesis that generates the rhythm in melatonin production (Ho et al., 2003). A direct photic stimulation of a pineal gland in explant culture results in suppression of p-ERK and a simultaneous decrease in melatonin precursor, suggesting that p-ERK connects neural stimulation to the production of melatonin (Ho et al., 2003). Several conflicting reports exist regarding whether a similar role for ERK in the chick pineal gland exists (Hayashi, Sanada, & Fukada, 2001; Sanada, Hayashi, Harada, Okano, & Fukada, 2000; Yadav et al., 2003). However, in the chick retina, ERK is rhythmically phosphorylated with its peak during subjective night, and rhythmic ERK regulates the rhythmic expression of L-type voltage-gated calcium channels that facilitate the rhythmic production of melatonin in photoreceptor cells (Ko et al., 2001). Together, these data indicate that the ERK pathway acts as a vital link between the circadian oscillator and overt rhythms in melatonin production in several tissue types.
In the mouse hippocampus, ERK appears to function in a circadian output pathway to modulate memory storage (Eckel-Mahan et al., 2008; Luo, Phan, Yang, Garelick, & Storm, 2013; Phan, Chan, Sindreu, Eckel-Mahan, & Storm, 2011; Shimizu, Phan, Mansuy, & Storm, 2007). In the hippocampus, p-ERK cycles with an endogenous rhythm peaking during subjective day, the time of day when mice are typically sleeping (Eckel-Mahan et al., 2008), and these rhythms are dependent on an intact SCN (Phan et al., 2011). Disruption of p-ERK rhythms prevents the consolidation, or storage, of long-term memories (Eckel-Mahan et al., 2008; Phan et al., 2011; Shimizu et al., 2007). Interestingly, p-ERK activity was higher during rapid eye movement sleep (Luo et al., 2013), an important stage in the sleep cycle for memory consolidation (Louie & Wilson, 2001; Poe, Nitz, McNaughton, & Barnes, 2000), further implicating ERK in the mechanism for memory consolidation.
Abundant evidence also exists in model organisms to understand how ERK functions in circadian output pathways. For example, the neurofibromatosis-1 (nf-1) gene in Drosophila encodes Ras GTPase, whose mutation leads to hyperactivation of rolled MAPK, the ERK homolog in Drosophila (Williams, Su, Bernards, Field, & Sehgal, 2001). In nf mutants, locomotor rhythms -1 are abolished in DD, but rhythms of circadian oscillator components are normal, suggesting that mutation of nf-1 disrupts a circadian output pathway downstream of the circadian oscillator (Williams et al., 2001). Additionally, staining of brains from flies kept in LD cycles revealed a rhythm in ERK activation that colocalized with pigment-dispersing factor (PDF), a neuropep-tide responsible for activity rhythms in the fly brain (Williams et al., 2001). Based on the colocalization of p-ERK with PDF, it was hypothesized that Ras/ERK acts downstream of PDF to regulate rhythmic behavior (Williams et al., 2001).
The ERK-related MAPKs in Neurospora, called MAK-1 and MAK-2, are rhythmically activated in constant conditions by the circadian clock (Bennett et al., 2013). As is a characteristic of function within an output pathway, mutation of these MAPKs did not disrupt the endogenous rhythmicity of oscillator components (Bennett et al., 2013). Transcriptional profiling from rhythmic cultures of either wild-type or Δmak-1 strains during the subjective morning identified potential target genes of MAK-1 (Bennett et al., 2013). Around 28% of the 517 putative MAK-1 targets were predicted to be CCGs, providing novel insights into the proportion of target genes that are rhythmically expressed from the circadian activity of an MAPK (Bennett et al., 2013).
6.2. p38 MAPK pathway
There are no data demonstrating a role for p38 MAPK in circadian output pathways in mammals, but model organisms have once again proved their usefulness in studying p38. The OS-2 MAPK in Neurospora, the p38 homolog, is rhythmically phosphorylated in constant conditions, and mutation of os-2 had no effect on the endogenous rhythms of the circadian oscillator, demonstrating that the OS-2 MAPK functions in circadian output (Vitalini et al., 2007). The OS MAPK pathway is a stress response pathway that facilitates an adaptive response after an osmotic shock (Noguchi et al., 2007). The endogenous rhythm of OS-2 MAPK activity peaks during the subjective morning, a time when the organism endures stress from the daily temperature increase and the onset of sunlight. Consistent with the prediction that circadian activation of OS-2 in early subjective morning prepares the organism to anticipate daily environmental stresses, the tissue challenged with an osmotic stress during the subjective morning mounted a more robust adaptive response compared to the tissue treated during the subjective night (Lamb et al., 2011). Presumably, this time of day difference is mediated via CCGs that are regulated by the OS-2 MAPK (Noguchi et al., 2007; Watanabe et al., 2007). An analysis of the downstream transcription factor ASL-1, the homolog to mammalian ATF-1, revealed that the rhythmicity of several CCGs induced by OS-2 activation required ASL-1 for endogenous circadian rhythmicity (Lamb et al., 2012). These data demonstrate that the circadian clock can utilize stress-responsive MAPK pathways to control rhythms in downstream targets that help to prepare the organism for predictable daily stress.
6.3. JNK MAPK pathway
To date, there is no evidence to demonstrate a role for JNK MAPK in circadian output pathways. However, JNK is rhythmically activated in rat-1 peripheral cells, suggesting that JNK may act within an output pathway in certain tissues.
6.4. Future directions
As a signaling molecule, the MAPK relies on interactions with a network of downstream components to elicit a biological effect. The components downstream of the MAPK provide the opportunity to integrate signals from other MAPK signaling pathways or, conversely, to confer signaling specificity. In an interesting example, both MSK-1 and RSK-1 are effector kinases that are phosphorylated by ERK in response to photic stimulation in the SCN; however, only RSK-1 is rhythmically phosphorylated as a consequence of the circadian rhythm in ERK activation. In Neurospora, the transcription factor ASL-1 is important for the rhythmicity of downstream CCGs, but it is not yet known if the transcription factor itself is rhythmically phosphorylated. It is presumed that rhythmic MAPK activation will lead to rhythms in downstream effectors to produce rhythmic transcriptional/translational activation of target genes. Additionally, the circadian clock may regulate the expression of effector genes downstream of an MAPK to reinforce the rhythmic expression of target genes. Indeed, transcriptional profiling has identified MAPK effector genes, such as MAPKAPK2, as CCGs (Menger et al., 2007). Surprisingly, no role for the AP-1 transcriptional activating complex, the primary downstream component of the JNK pathway, has been described despite findings of rhythms in JNK activity. Additionally, ERK appears to signal through mTOR in mouse SCN to regulate translation, which may generate or reinforce rhythmic gene expression. Thus, rhythmic control of downstream effectors that regulate the transcription and translation of target genes constitutes a key area for future research.
As demonstrated by the OS MAPK pathway in Neurospora, the circadian rhythms in MAPK activation provide a mechanism for the organism to defend against certain stresses during specific times of the day. Stress response, especially in p38 and JNK pathways, is a primary function of MAPK pathways and may prove vital to the function of some tissues. For example, the p38 MAPK is essential for mediating an inflammatory response in immune cells. It is known that self-sustaining circadian oscillators exist in some immune cells (Boivin et al., 2003; Keller et al., 2009), and therefore, it is likely that a circadian rhythm in p38 MAPK activity would have a profound effect in these cells. Additionally, ERK activity has been shown to confer resistance to glutamate-induced neurotoxicity (Karmarkar, Bottum, Krager, & Tischkau, 2011). This suggests that rhythms in ERK activity may confer an increased resistance to naturally occurring incidences of neu-rotoxicity, such as stroke or brain damage, at certain times of the day. MAPKs have also been shown to be important regulators of the cell cycle and apoptosis in response to DNA damage, leading to the hypothesis that rhythmic MAPK activation may play a key role in cell survival throughout the circadian cycle.
7. CONCLUDING REMARKS
Through their ability to relay biochemical signals within the cell in response to stimuli, MAPK pathways are important components of the circadian clock. Phosphorylation of clock proteins by all three MAPK subfamilies is apparent in diverse eukaryotic organisms; however, the biological significance of clock protein phosphorylation by MAPKs is still poorly understood. While the role of MAPKs in circadian input pathways is firmly established, a role for these signal-transduction pathways in circadian output is only beginning to emerge. The activity of MAPKs is frequently regulated by the clock in diverse tissues, but this rhythmic activation has not been connected to known downstream signaling components. The rhythmic interaction of MAPKs with downstream components will expand the understanding of MAPKs’ ability to rhythmically regulate gene expression and translational control, and this information would be useful to help clarify the role of MAPKs in controlling CCG expression. Finally, as the role of the circadian clocks is more thoroughly understood in peripheral tissues, MAPKs will likely be contributors to circadian rhythms due to their ability to assume unique functions in disparate tissues. Despite all of the questions that remain unanswered, studies of the link between MAPK and the circadian clock system are moving forward, with model systems leading the way. Unraveling the mechanistic connections between the clock and MAPK pathways in model systems, such as fungi, will undoubtedly provide the foundation for solving the problem in complex mammalian systems. Once achieved, we will be in a position to develop novel therapies to treat diseases shared by these two pathways, including regulation of cell growth mechanisms, immune disorders, heart disease, and neurodegenerative disorders (Cuenda & Rousseau, 2007; Fu & Lee, 2003; Han & Sun, 2007).
References
- Akashi M, Hayasaka N, Yamazaki S, Node K. Mitogen-activated protein kinase is a functional component of the autonomous circadian system in the sup-rachiasmatic nucleus. The Journal of Neuroscience. 2008;28(18):4619–4623. doi: 10.1523/JNEUROSCI.3410-07.2008. http://dx.doi.org/10.1523/JNEUROSCI.3410-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi M, Nishida E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes & Development. 2000;14(6):645–649. http://dx.doi.org/10.1101/gad.14.6.645. [PMC free article] [PubMed] [Google Scholar]
- Anjum R, Blenis J. The RSK family of kinases: Emerging roles in cellular signalling. Nature Reviews Molecular Cell Biology. 2008;9(10):747–758. doi: 10.1038/nrm2509. http://dx.doi.org/10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- Antoun G, Cannon PB, Cheng HYM. Regulation of MAPK/ERK signaling and photic entrainment of the suprachiasmatic nucleus circadian clock by Raf kinase inhibitor protein. The Journal of Neuroscience. 2012;32(14):4867–4877. doi: 10.1523/JNEUROSCI.5650-11.2012. http://dx.doi.org/10.1523/jneurosci.5650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur J, Fong AL, Dwyer JM, Davare M, Reese E, Obrietan K. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. The Journal of Neuroscience. 2004;24(18):4324–4332. doi: 10.1523/JNEUROSCI.5227-03.2004. http://dx.doi.org/10.1523/jneurosci.5227-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley MC, Davis T, Dix MC, Rokicki MJ, Kipling D. Rapid synthesis of VX-745: p38 MAP kinase inhibition in Werner syndrome cells. Bioorganic & Medicinal Chemistry Letters. 2007;17(18):5107–5110. doi: 10.1016/j.bmcl.2007.07.016. http://dx.doi.org/10.1016/j.bmcl.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: An update. Biochemical Journal. 2003;371(Pt 1):199–204. doi: 10.1042/BJ20021535. http://dx.doi.org/10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured rat-1 fibroblasts. Current Biology. 2000;10(20):1291–1294. doi: 10.1016/s0960-9822(00)00758-2. http://dx.doi.org/10.1016/S0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- Barr RK, Bogoyevitch MA. The c-Jun N-terminal protein kinase family of mitogen-activated protein kinases (JNK MAPKs) The International Journal of Biochemistry & Cell Biology. 2001;33(11):1047–1063. doi: 10.1016/s1357-2725(01)00093-0. http://dx.doi.org/10.1016/S1357-2725(01)00093-0. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nature Reviews Genetics. 2005;6(7):544–556. doi: 10.1038/nrg1633. http://dx.doi.org/10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LD, Beremand P, Thomas TL, Bell-Pedersen D. Circadian activation of the mitogen-activated protein kinase MAK-1 facilitates rhythms in clock-controlled genes in Neurospora crassa. Eukaryotic Cell. 2013;12(1):59–69. doi: 10.1128/EC.00207-12. http://dx.doi.org/10.1128/ec.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13681–13686. doi: 10.1073/pnas.251194298. http://dx.doi.org/10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch MA. The isoform-specific functions of the c-Jun N-terminal Kinases (JNKs): Differences revealed by gene targeting. Bioessays. 2006;28(9):923–934. doi: 10.1002/bies.20458. http://dx.doi.org/10.1002/bies.20458. [DOI] [PubMed] [Google Scholar]
- Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102(12):4143–4145. doi: 10.1182/blood-2003-03-0779. http://dx.doi.org/10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- Bozek K, Rosahl AL, Gaub S, Lorenzen S, Herzel H. Circadian transcription in liver. Biosystems. 2010;102(1):61–69. doi: 10.1016/j.biosystems.2010.07.010. http://dx.doi.org/10.1016/j.biosystems.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Bray MS, Shaw CA, Moore MWS, Garcia RAP, Zanquetta MM, Durgan DJ. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. American Journal of Physiology - Heart and Circulatory Physiology. 2008;294(2):H1036–H1047. doi: 10.1152/ajpheart.01291.2007. http://dx.doi.org/10.1152/ajpheart.01291.2007. [DOI] [PubMed] [Google Scholar]
- Butcher GQ, Doner J, Dziema H, Collamore M, Burgoon PW, Obrietan K. The p42/44 mitogen-activated protein kinase pathway couples photic input to circadian clock entrainment. The Journal of Biological Chemistry. 2002;277(33):29519–29525. doi: 10.1074/jbc.M203301200. http://dx.doi.org/10.1074/jbc.M203301200. [DOI] [PubMed] [Google Scholar]
- Butcher GQ, Lee B, Cheng HY, Obrietan K. Light stimulates MSK1 activation in the suprachiasmatic nucleus via a PACAP-ERK/MAP kinase-dependent mechanism. The Journal of Neuroscience. 2005;25(22):5305–5313. doi: 10.1523/JNEUROSCI.4361-04.2005. http://dx.doi.org/10.1523/JNEUROSCI.4361-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher GQ, Lee B, Hsieh F, Obrietan K. Light- and clock-dependent regulation of ribosomal S6 kinase activity in the suprachiasmatic nucleus. European Journal of Neuroscience. 2004;19(4):907–915. doi: 10.1111/j.0953-816x.2004.03155.x. http://dx.doi.org/10.1111/j.0953-816X.2004.03155.x. [DOI] [PubMed] [Google Scholar]
- Cahill GM. Clock mechanisms in zebrafish. Cell and Tissue Research. 2002;309(1):27–34. doi: 10.1007/s00441-002-0570-7. http://dx.doi.org/10.1007/s00441-002-0570-7. [DOI] [PubMed] [Google Scholar]
- Cao R, Anderson FE, Jung YJ, Dziema H, Obrietan K. Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience. 2011;181:79–88. doi: 10.1016/j.neuroscience.2011.03.005. http://dx.doi.org/10.1016/j.neuroscience.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Butcher GQ, Karelina K, Arthur JS, Obrietan K. Mitogen- and stress-activated protein kinase 1 modulates photic entrainment of the suprachiasmatic circadian clock. European Journal of Neuroscience. 2013;37(1):130–140. doi: 10.1111/ejn.12028. http://dx.doi.org/10.1111/ejn.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Lee B, Cho HY, Saklayen S, Obrietan K. Photic regulation of the mTOR signaling pathway in the suprachiasmatic circadian clock. Molecular and Cellular Neuroscience. 2008;38(3):312–324. doi: 10.1016/j.mcn.2008.03.005. http://dx.doi.org/10.1016/j.mcn.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Li A, Cho HY, Lee B, Obrietan K. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. The Journal of Neuroscience. 2010;30(18):6302–6314. doi: 10.1523/JNEUROSCI.5482-09.2010. http://dx.doi.org/10.1523/JNEUROSCI.5482-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N, Pando MP, Thompson CL, Pinchak AB, Selby CP, Gutierrez L. Light induction of a vertebrate clock gene involves signaling through blue-light receptors and MAP kinases. Current Biology. 2002;12(10):844–848. doi: 10.1016/s0960-9822(02)00835-7. http://dx.doi.org/10.1016/S0960-9822(02)00835-7. [DOI] [PubMed] [Google Scholar]
- Chansard M, Molyneux P, Nomura K, Harrington ME, Fukuhara C. c-Jun N-terminal kinase inhibitor SP600125 modulates the period of mammalian circadian rhythms. Neuroscience. 2007;145(3):812–823. doi: 10.1016/j.neuroscience.2006.12.037. http://dx.doi.org/10.1016/j.neuroscience.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, DeMay BS, Gladfelter AS, Dunlap JC, Loros JJ. Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(38):16715–16720. doi: 10.1073/pnas.1011190107. http://dx.doi.org/10.1073/pnas.1011190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HYM, Dziema H, Papp J, Mathur DP, Koletar M, Ralph MR. The molecular gatekeeper Dexras1 sculpts the photic responsiveness of the mammalian circadian clock. The Journal of Neuroscience. 2006;26(50):12984–12995. doi: 10.1523/JNEUROSCI.4253-06.2006. http://dx.doi.org/10.1523/jneurosci.4253-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HYM, Obrietan K, Cain SW, Lee BY, Agostino PV, Joza NA. Dexras1 potentiates photic and suppresses nonphotic responses of the circadian clock. Neuron. 2004;43(5):715–728. doi: 10.1016/j.neuron.2004.08.021. http://dx.doi.org/10.1016/j.neuron.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Cheng H-YM, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–829. doi: 10.1016/j.neuron.2007.05.017. http://dx.doi.org/10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chik CL, Mackova M, Price D, Ho AK. Adrenergic regulation and diurnal rhythm of p38 mitogen-activated protein kinase phosphorylation in the rat pineal gland. Endocrinology. 2004;145(11):5194–5201. doi: 10.1210/en.2004-0864. http://dx.doi.org/10.1210/en.2004-0864. [DOI] [PubMed] [Google Scholar]
- Coogan AN, Piggins HD. Circadian and photic regulation of phosphorylation of ERK1/2 and Elk-1 in the suprachiasmatic nuclei of the syrian hamster. The Journal of Neuroscience. 2003;23(7):3085–3093. doi: 10.1523/JNEUROSCI.23-07-03085.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coogan AN, Piggins HD. MAP kinases in the mammalian circadian system—Key regulators of clock function. Journal of Neurochemistry. 2004;90(4):769–775. doi: 10.1111/j.1471-4159.2004.02554.x. http://dx.doi.org/10.1111/j.1471-4159.2004.02554.x. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. p38 MAP-Kinases pathway regulation, function and role in human diseases. Biochimica et Biophysica Acta—Molecular Cell Research. 2007;1773(8):1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. http://dx.doi.org/10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Daan S, Pittendrigh CS. A functional analysis of circadian pacemakers in nocturnal rodents. IV. Entrainment: Pacemaker as clock. Journal of Comparative Physiology. 1976;106:253–266. [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. http://dx.doi.org/10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Davis S, Vanhoutte P, Pagès C, Caboche J, Laroche S. The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. The Journal of Neuroscience. 2000;20(12):4563–4572. doi: 10.1523/JNEUROSCI.20-12-04563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula RM, Lamb TM, Bennett L, Bell-Pedersen D. A connection between MAPK pathways and circadian clocks. Cell Cycle. 2008;7(17):2630–2634. doi: 10.4161/cc.7.17.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annual Review of Physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. http://dx.doi.org/10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96(2):271–290. doi: 10.1016/s0092-8674(00)80566-8. http://dx.doi.org/10.1016/S0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, Liu Y, Crosthwaite SK. Eukaryotic circadian systems: Cycles in common. Genes to Cells. 1999;4(1):1–10. doi: 10.1046/j.1365-2443.1999.00239.x. http://dx.doi.org/10.1046/j.1365-2443.1999.00239.x. [DOI] [PubMed] [Google Scholar]
- Dziema H, Oatis B, Butcher GQ, Yates R, Hoyt KR, Obrietan K. The ERK/MAP kinase pathway couples light to immediate-early gene expression in the sup-rachiasmatic nucleus. European Journal of Neuroscience. 2003;17(8):1617–1627. doi: 10.1046/j.1460-9568.2003.02592.x. http://dx.doi.org/10.1046/j.1460-9568.2003.02592.x. [DOI] [PubMed] [Google Scholar]
- Dziema H, Obrietan K. PACAP potentiates L-type calcium channel conductance in suprachiasmatic nucleus neurons by activating the MAPK pathway. Journal of Neurophysiology. 2002;88(3):1374–1386. doi: 10.1152/jn.2002.88.3.1374. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GCK, Scheiner ZS. Circadian oscillation of hippocampal MAPK activity and cAMP: Implications for memory persistence. Nature Neuroscience. 2008;11(9):1074–1082. doi: 10.1038/nn.2174. http://dx.doi.org/10.1038/nn.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG. A small molecule-kinase interaction map for clinical kinase inhibitors. Nature Biotechnology. 2005;23(3):329–336. doi: 10.1038/nbt1068. http://dx.doi.org/10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Fu L, Lee CC. The circadian clock: Pacemaker and tumour suppressor. Nature Reviews Cancer. 2003;3(5):350–361. doi: 10.1038/nrc1072. http://dx.doi.org/10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- Gazel A, Banno T, Walsh R, Blumenberg M. Inhibition of JNK promotes differentiation of epidermal keratinocytes. The Journal of Biological Chemistry. 2006;281(29):20530–20541. doi: 10.1074/jbc.M602712200. http://dx.doi.org/10.1074/jbc.M602712200. [DOI] [PubMed] [Google Scholar]
- Gazel A, Nijhawan RI, Walsh R, Blumenberg M. Transcriptional profiling defines the roles of ERK and p38 kinases in epidermal keratinocytes. Journal of Cellular Physiology. 2008;215(2):292–308. doi: 10.1002/jcp.21394. http://dx.doi.org/10.1002/jcp.21394. [DOI] [PubMed] [Google Scholar]
- Ginty DD, Kornhauser JM, Thompson MA, Bading H, Mayo KE, Takahashi JS. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260(5105):238–241. doi: 10.1126/science.8097062. http://dx.doi.org/10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- Golden SS, Ishiura M, Johnson CH, Kondo T. Cyanobacterial circadian rhythms. Annual Review of Plant Biology. 1997;48(1):327–354. doi: 10.1146/annurev.arplant.48.1.327. http://dx.doi.org/10.1146/annurev.arplant.48.1.327. [DOI] [PubMed] [Google Scholar]
- Hagiwara D, Asano Y, Marui J, Yoshimi A, Mizuno T, Abe K. Transcriptional profiling for Aspergillus nidulans HogA MAPK signaling pathway in response to fludioxonil and osmotic stress. Fungal Genetics and Biology. 2009;46(11):868–878. doi: 10.1016/j.fgb.2009.07.003. http://dx.doi.org/10.1016/j.fgb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Hainich EC, Pizzio GA, Golombek DA. Constitutive activation of the ERK-MAPK pathway in the suprachiasmatic nuclei inhibits circadian resetting. FEBS Letters. 2006;580(28–29):6665–6668. doi: 10.1016/j.febslet.2006.11.019. http://dx.doi.org/10.1016/j.febslet.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Han J, Sun P. The pathways to tumor suppression via route p38. Trends in Biochemical Sciences. 2007;32(8):364–371. doi: 10.1016/j.tibs.2007.06.007. http://dx.doi.org/10.1016/j.tibs.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Hardin PE. The circadian timekeeping system of Drosophila. Current Biology. 2005;15(17):R714–R722. doi: 10.1016/j.cub.2005.08.019. http://dx.doi.org/10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Cahill GM. Regulation of the circadian oscillator in Xenopus retinal photoreceptors by protein kinases sensitive to the stress-activated protein kinase inhibitor, SB 203580. The Journal of Biological Chemistry. 2004;279(21):22738–22746. doi: 10.1074/jbc.M401389200. http://dx.doi.org/10.1074/jbc.M401389200. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Sanada K, Fukada Y. Circadian and photic regulation of MAP kinase by Ras- and protein phosphatase-dependent pathways in the chick pineal gland. FEBS Letters. 2001;491(1–2):71–75. doi: 10.1016/s0014-5793(01)02153-6. http://dx.doi.org/10.1016/S0014-5793(01)02153-6. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Sanada K, Hirota T, Shimizu F, Fukada Y. p38 mitogen-activated protein kinase regulates oscillation of chick pineal circadian clock. The Journal of Biological Chemistry. 2003;278(27):25166–25171. doi: 10.1074/jbc.M212726200. http://dx.doi.org/10.1074/jbc.M212726200. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: Quarrel and harmony among siblings. Journal of Cell Science. 2004;117(25):5965–5973. doi: 10.1242/jcs.01589. http://dx.doi.org/10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Ho AK, Mackova M, Price L, Chik CL. Diurnal variation in p42/44 mitogen-activated protein kinase in the rat pineal gland. Molecular and Cellular Endocri-nology. 2003;208(1–2):23–30. doi: 10.1016/s0303-7207(03)00260-0. http://dx.doi.org/10.1016/S0303-7207(03)00260-0. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Elliott JA, Foster R. Entrainment of circadian programs. Chro-nobiology International. 2003;20(5):741–774. doi: 10.1081/cbi-120024211. http://dx.doi.org/10.1081/cbi-120024211. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. http://dx.doi.org/10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- Karmarkar SW, Bottum KM, Krager SL, Tischkau SA. ERK/MAPK is essential for endogenous neuroprotection in SCN2.2 cells. PLoS One. 2011;6(8):e23493. doi: 10.1371/journal.pone.0023493. http://dx.doi.org/10.1371/journal.pone.0023493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD. A circadian clock in macrophages controls inflammatory immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21407–21412. doi: 10.1073/pnas.0906361106. http://dx.doi.org/10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Ie. Cell. 1998;94(1):97–107. doi: 10.1016/s0092-8674(00)81225-8. http://dx.doi.org/10.1016/S0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Ko GYP, Ko ML, Dryer SE. Circadian regulation of cGMP-gated cationic channels of chick retinal cones: Erk MAP kinase and Ca2+/calmodulin-dependent protein kinase II. Neuron. 2001;29(1):255–266. doi: 10.1016/s0896-6273(01)00195-7. http://dx.doi.org/10.1016/S0896-6273(01)00195-7. [DOI] [PubMed] [Google Scholar]
- Ko ML, Shi L, Ko GY. Circadian controls outweigh acute illumination effects on the activity of extracellular signal-regulated kinase (ERK) in the retina. Neuroscience Letters. 2009;451(1):74–78. doi: 10.1016/j.neulet.2008.12.025. http://dx.doi.org/10.1016/j.neulet.2008.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko ML, Shi L, Tsai JY, Young ME, Neuendorff N, Earnest DJ. Cardiac-specific mutation of clock alters the quantitative measurements of physical activities without changing behavioral circadian rhythms. Journal of Biological Rhythms. 2011;26(5):412–422. doi: 10.1177/0748730411414170. http://dx.doi.org/10.1177/0748730411414170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Human Molecular Genetics. 2006;15(suppl 2):R271–R277. doi: 10.1093/hmg/ddl207. http://dx.doi.org/10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. http://dx.doi.org/10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nature Reviews Molecular Cell Biology. 2005;6(11):827–837. doi: 10.1038/nrm1743. http://dx.doi.org/10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Finch KE, Bell-Pedersen D. The Neurospora crassa OS MAPK pathway-activated transcription factor ASL-1 contributes to circadian rhythms in pathway responsive clock-controlled genes. Fungal Genetics and Biology. 2012;49(2):180–188. doi: 10.1016/j.fgb.2011.12.006. http://dx.doi.org/10.1016/j.fgb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TM, Goldsmith CS, Bennett L, Finch KE, Bell-Pedersen D. Direct transcriptional control of a p38 MAPK pathway by the circadian clock in Neurospora crassa. PLoS One. 2011;6(11):e27149. doi: 10.1371/journal.pone.0027149. http://dx.doi.org/10.1371/journal.pone.0027149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Dunlap JC, Loros JJ. Interconnected feedback loops in the Neuros-pora circadian system. Science. 2000;289(5476):107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]
- Lee HS, Nelms JL, Nguyen M, Silver R, Lehman MN. The eye is necessary for a circadian rhythm in the suprachiasmatic nucleus. Nature Neuroscience. 2003;6(2):111–112. doi: 10.1038/nn1006. http://dx.doi.org/10.1038/nn1006. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35(4):605–623. doi: 10.1016/s0896-6273(02)00828-0. http://dx.doi.org/10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Louie K, Wilson MA. Temporally structured replay of awake hippocampal ensemble activity during rapid eye movement sleep. Neuron. 2001;29(1):145–156. doi: 10.1016/s0896-6273(01)00186-6. http://dx.doi.org/10.1016/S0896-6273(01)00186-6. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288(5465):483–491. doi: 10.1126/science.288.5465.483. http://dx.doi.org/10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Phan TX, Yang Y, Garelick MG, Storm DR. Increases in cAMP, MAPK activity, and CREB phosphorylation during REM sleep: Implications for REM sleep and memory consolidation. The Journal of Neuroscience. 2013;33(15):6460–6468. doi: 10.1523/JNEUROSCI.5018-12.2013. http://dx.doi.org/10.1523/jneurosci.5018-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1 doi: 10.7554/eLife.00011. http://dx.doi.org/10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menger GJ, Allen GC, Neuendorff N, Nahm SS, Thomas TL, Cassone VM. Circadian profiling of the transcriptome in NIH/3T3 fibroblasts: Comparison with rhythmic gene expression in SCN2.2 cells and the rat SCN. Physiological Genomics. 2007;29(3):280–289. doi: 10.1152/physiolgenomics.00199.2006. http://dx.doi.org/10.1152/physiolgenomics.00199.2006. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annual Review of Neuroscience. 2012;35(1):445–462. doi: 10.1146/annurev-neuro-060909-153128. http://dx.doi.org/10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annual Review of Cell and Developmental Biology. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. http://dx.doi.org/10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- Mrosovsky N. Locomotor activity and non-photic influences on circadian clocks. Biological Reviews. 1996;71(3):343–372. doi: 10.1111/j.1469-185x.1996.tb01278.x. http://dx.doi.org/10.1111/j.1469-185X.1996.tb01278.x. [DOI] [PubMed] [Google Scholar]
- Nakaya M, Sanada K, Fukada Y. Spatial and temporal regulation of mitogen-activated protein kinase phosphorylation in the mouse suprachiasmatic nucleus. Biochemical and Biophysical Research Communications. 2003;305(3):494–501. doi: 10.1016/s0006-291x(03)00791-5. http://dx.doi.org/10.1016/S0006-291X(03)00791-5. [DOI] [PubMed] [Google Scholar]
- Noguchi R, Banno S, Ichikawa R, Fukumori F, Ichiishi A, Kimura M. Identification of OS-2 MAP kinase-dependent genes induced in response to osmotic stress, antifungal agent fludioxonil, and heat shock in Neurospora crassa. Fungal Genetics and Biology. 2007;44(3):208–218. doi: 10.1016/j.fgb.2006.08.003. http://dx.doi.org/10.1016/j.fgb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. The Journal of Biological Chemistry. 1999;274(25):17748–17756. doi: 10.1074/jbc.274.25.17748. http://dx.doi.org/10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nature Neuroscience. 1998;1(8):693–700. doi: 10.1038/3695. http://dx.doi.org/10.1038/3695. [DOI] [PubMed] [Google Scholar]
- O’Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes & Development. 1998;12(18):2874–2886. doi: 10.1101/gad.12.18.2874. http://dx.doi.org/10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26(22):3203–3213. doi: 10.1038/sj.onc.1210412. http://dx.doi.org/10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- Petrzilka S, Taraborrelli C, Cavadini G, Fontana A, Birchler T. Clock gene modulation by TNF-alpha depends on calcium and p38 MAP kinase signaling. Journal of Biological Rhythms. 2009;24(4):283–294. doi: 10.1177/0748730409336579. http://dx.doi.org/10.1177/0748730409336579. [DOI] [PubMed] [Google Scholar]
- Phan TH, Chan GCK, Sindreu CB, Eckel-Mahan KL, Storm DR. The diurnal oscillation of MAP (mitogen-activated protein) kinase and adenylyl cyclase activities in the hippocampus depends on the suprachiasmatic nucleus. The Journal of Neu-roscience. 2011;31(29):10640–10647. doi: 10.1523/JNEUROSCI.6535-10.2011. http://dx.doi.org/10.1523/jneurosci.6535-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzio GA, Hainich EC, Ferreyra GA, Coso OA, Golombek DA. Circadian and photic regulation of ERK, JNK and p38 in the hamster SCN. Neuroreport. 2003;14(11):1417–1419. doi: 10.1097/00001756-200308060-00002. http://dx.doi.org/10.1097/01.wnr.0000082025.91120.69. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278(5343):1632–1635. doi: 10.1126/science.278.5343.1632. http://dx.doi.org/10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Poe GR, Nitz DA, McNaughton BL, Barnes CA. Experience-dependent phase-reversal of hippocampal neuron firing during REM sleep. Brain Research. 2000;855(1):176–180. doi: 10.1016/s0006-8993(99)02310-0. http://dx.doi.org/10.1016/S0006-8993(99)02310-0. [DOI] [PubMed] [Google Scholar]
- Pollard LC, Choy EH, Gonzalez J, Khoshaba B, Scott DL. Fatigue in rheumatoid arthritis reflects pain, not disease activity. Rheumatology. 2006;45(7):885–889. doi: 10.1093/rheumatology/kel021. http://dx.doi.org/10.1093/rheumatology/kel021. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94(1):83–95. doi: 10.1016/s0092-8674(00)81224-6. http://dx.doi.org/10.1016/S0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. http://dx.doi.org/10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287(5454):873–880. doi: 10.1126/science.287.5454.873. http://dx.doi.org/10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M. Molecular circadian oscillators: An alternative hypothesis. Journal of Biological Rhythms. 1998;13(2):167–179. doi: 10.1177/074873098129000011. http://dx.doi.org/10.1177/074873098129000011. [DOI] [PubMed] [Google Scholar]
- Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiology and Molecular Biology Reviews. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. http://dx.doi.org/10.1128/mmbr.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K, Harada Y, Sakai M, Todo T, Fukada Y. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes to Cells. 2004;9(8):697–708. doi: 10.1111/j.1356-9597.2004.00758.x. http://dx.doi.org/10.1111/j.1356-9597.2004.00758.x. [DOI] [PubMed] [Google Scholar]
- Sanada K, Hayashi Y, Harada Y, Okano T, Fukada Y. Role of circadian activation of mitogen-activated protein kinase in chick pineal clock oscillation. The Journal of Neuroscience. 2000;20(3):986–991. doi: 10.1523/JNEUROSCI.20-03-00986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K, Okano T, Fukada Y. Mitogen-activated protein kinase phosphor-ylates and negatively regulates basic helix-loop-helix-PAS transcription factor BMAL1. The Journal of Biological Chemistry. 2002;277(1):267–271. doi: 10.1074/jbc.M107850200. http://dx.doi.org/10.1074/jbc.M107850200. [DOI] [PubMed] [Google Scholar]
- Schmidt TM, Chen SK, Hattar S. Intrinsically photosensitive retinal ganglion cells: Many subtypes, diverse functions. Trends in Neurosciences. 2011;34(11):572–580. doi: 10.1016/j.tins.2011.07.001. http://dx.doi.org/10.1016/j.tins.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurov IL, Hepworth TJ, Hastings MH. Dopaminergic signalling in the rodent neonatal suprachiasmatic nucleus identifies a role for protein kinase A and mitogen-activated protein kinase in circadian entrainment. European Journal of Neurosci-ence. 2002;15(2):223–232. doi: 10.1046/j.0953-816x.2001.01848.x. http://dx.doi.org/10.1046/j.0953-816x.2001.01848.x. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Okada M, Nagai K, Fukada Y. Suprachiasmatic nucleus circadian oscillatory protein, a novel binding partner of K-Ras in the membrane rafts, negatively regulates MAPK pathway. The Journal of Biological Chemistry. 2003;278(17):14920–14925. doi: 10.1074/jbc.M213214200. http://dx.doi.org/10.1074/jbc.M213214200. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Okada M, Takano A, Nagai K. SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Letters. 1999;458(3):363–369. doi: 10.1016/s0014-5793(99)01190-4. http://dx.doi.org/10.1016/S0014-5793(99)01190-4. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Phan T, Mansuy IM, Storm DR. Proteolytic degradation of SCOP in the hippocampus contributes to activation of MAP kinase and memory. Cell. 2007;128(6):1219–1229. doi: 10.1016/j.cell.2006.12.047. http://dx.doi.org/10.1016/j.cell.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Sancar G, Dekhang R, Sullivan CM, Li S, Tag AG. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for Neurospora white collar complex. Eukaryotic Cell. 2010;9(10):1549–1556. doi: 10.1128/EC.00154-10. http://dx.doi.org/10.1128/ec.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs DR, Sherman ML, Michie H, Arthur KA, Imamura K, Wilmore D. Recombinant human tumor necrosis factor administered as a 24-hour intravenous infusion: A phase I and pharmacologic study. Journal of the National Cancer Institute. 1988;80(13):1039–1044. doi: 10.1093/jnci/80.13.1039. http://dx.doi.org/10.1093/jnci/80.13.1039. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. http://dx.doi.org/10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Tauber E, Last KS, Olive PJW, Kyriacou CP. Clock gene evolution and functional divergence. Journal of Biological Rhythms. 2004;19(5):445–458. doi: 10.1177/0748730404268775. http://dx.doi.org/10.1177/0748730404268775. [DOI] [PubMed] [Google Scholar]
- Tischkau SA, Mitchell JW, Tyan SH, Buchanan GF, Gillette MU. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. The Journal of Biological Chemistry. 2003;278(2):718–723. doi: 10.1074/jbc.M209241200. http://dx.doi.org/10.1074/jbc.M209241200. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Minami I, Kadotani H, Todo T, Nishida E. Circadian clock-controlled diurnal oscillation of Ras/ERK signaling in mouse liver. Proceedings of the Japan Academy Series B, Physical and Biological Sciences. 2013;89(1):59–65. doi: 10.2183/pjab.89.59. http://dx.doi.org/10.2183/pjab.89.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Osaki T, Yamasaki T, Shimomura T, Hata S, Horikawa K. Involvement of stress kinase mitogen-activated protein kinase kinase 7 in regulation of mammalian circadian clock. The Journal of Biological Chemistry. 2012;287(11):8318–8326. doi: 10.1074/jbc.M111.308908. http://dx.doi.org/10.1074/jbc.M111.308908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoutte P, Barnier JV, Guibert B, Pagès C, Besson MJ, Hipskind RA. Glutamate induces phosphorylation of Elk-1 and CREB, along with c-fos activation, via an extracellular signal-regulated kinase-dependent pathway in brain slices. Molecular and Cellular Biology. 1999;19(1):136–146. doi: 10.1128/mcb.19.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vgontzas AN, Zoumakis E, Lin HM, Bixler EO, Trakada G, Chrousos GP. Marked decrease in sleepiness in patients with sleep apnea by etanercept, a tumor necrosis factor-a antagonist. The Journal of Clinical Endocrinology and Metabolism. 2004;89(9):4409–4413. doi: 10.1210/jc.2003-031929. http://dx.doi.org/10.1210/jc.2003-031929. [DOI] [PubMed] [Google Scholar]
- Vitalini MW, de Paula RM, Goldsmith CS, Jones CA, Borkovich KA, Bell-Pedersen D. Circadian rhythmicity mediated by temporal regulation of the activity of p38 MAPK. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(46):18223–18228. doi: 10.1073/pnas.0704900104. http://dx.doi.org/10.1073/pnas.0704900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalini MW, de Paula RM, Park WD, Bell-Pedersen D. The rhythms of life: Circadian output pathways in Neurospora. Journal of Biological Rhythms. 2006;21(6):432–444. doi: 10.1177/0748730406294396. http://dx.doi.org/10.1177/0748730406294396. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metabolism. 2012;16(6):833–845. doi: 10.1016/j.cmet.2012.11.004. http://dx.doi.org/10.1016/j.cmet.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GK, Sehgal A. Signaling components that drive circadian rhythms. Current Opinion in Neurobiology. 2002;12(3):331–338. doi: 10.1016/s0959-4388(02)00324-0. http://dx.doi.org/10.1016/S0959-4388(02)00324-0. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Yamashita K, Ochiai N, Fukumori F, Ichiishi A, Kimura M. OS-2 mitogen activated protein kinase regulates the clock-controlled gene ccg-1 in Neurospora crassa. Bioscience, Biotechnology, and Biochemistry. 2007;71(11):2856–2859. doi: 10.1271/bbb.70410. http://dx.doi.org/10.1271/bbb.70410. [DOI] [PubMed] [Google Scholar]
- Weber F, Hung HC, Maurer C, Kay SA. Second messenger and Ras/MAPK signalling pathways regulate CLOCK/CYCLE-dependent transcription. Journal of Neurochemistry. 2006;98(1):248–257. doi: 10.1111/j.1471-4159.2006.03865.x. http://dx.doi.org/10.1111/j.1471-4159.2006.03865.x. [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends in Biochemical Sciences. 1998;23(12):481–485. doi: 10.1016/s0968-0004(98)01309-7. http://dx.doi.org/10.1016/S0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293(5538):2251–2256. doi: 10.1126/science.1063097. http://dx.doi.org/10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273(5277):959–963. doi: 10.1126/science.273.5277.959. http://dx.doi.org/10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- Yadav G, Straume M, Heath J, 3rd, Zatz M. Are changes in MAPK/ERK necessary or sufficient for entrainment in chick pineal cells? The Journal of Neuroscience. 2003;23(31):10021–10031. doi: 10.1523/JNEUROSCI.23-31-10021.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Yamanaka I, Koinuma S, Shigeyoshi Y, Uchiyama Y. Mini screening of kinase inhibitors affecting period-length of mammalian cellular circadian clock. Acta Histochemica et Cytochemica. 2009;42(3):89–93. doi: 10.1267/ahc.09015. http://dx.doi.org/10.1267/ahc.09015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitane H, Honma S, Imamura K, Nakajima H, Nishide SY, Ono D. JNK regulates the photic response of the mammalian circadian clock. EMBO Reports. 2012;13(5):455–461. doi: 10.1038/embor.2012.37. http://dx.doi.org/10.1038/embor.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Research. 2005;15(1):11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]