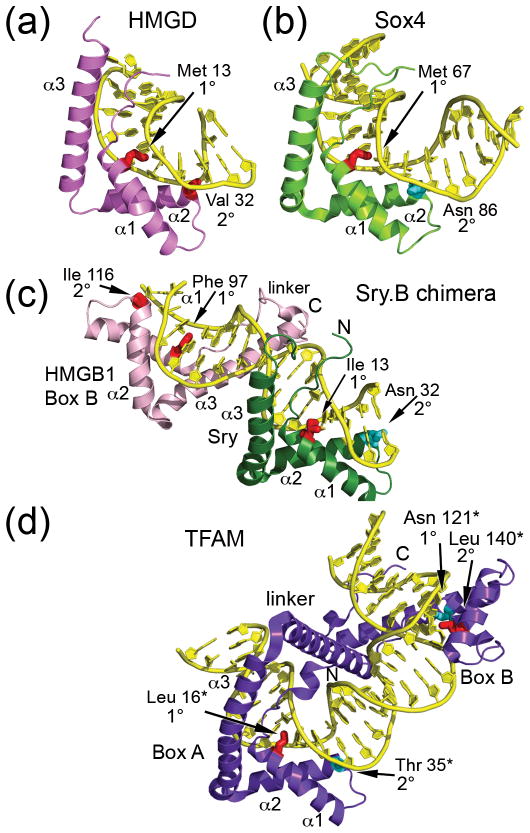
Figure 2. Structures of high mobility group (HMG)-box-DNA complexes.
(a) Non-sequence-specific HMGD bound to an unmodified DNA decamer (PDB ID 1QRV) [33]. (b) Sequence-specific Sox4 bound to a 16 base pair DNA oligomer from the Lama 1 gene (PDB ID 3U2B) [41]. The structure is orientated as if it was superimposed onto the HMG box of HMGD in panel (a). (c) Dual HMG box Sex determining region Y-HMGB1 Box B chimera (Sry.B) bound to 16 base pair DNA oligomer containing the cognate Sry binding site (PDB ID 2GZK) [29]. SRY is in green and the HMGB1 box B is in pink. (d) Native dual HMG box mitochondrial transcription factor A (TFAM) bound to a 28 base pair segment of the human mitochondrial light strand promoter (PDB ID 3TMM) [31]. The numbering for TFAM is based on the residual protein after loss of the 42 amino acid residue long mitochondrial localization sequence, indicated by a *. The structure is oriented as if box A was superimposed onto the SRY HMG box from the SRY.B chimera in panel (c). The non-polar intercalating residues are shown in red, and polar residues at the 1° or 2° intercalation sites are shown in cyan.