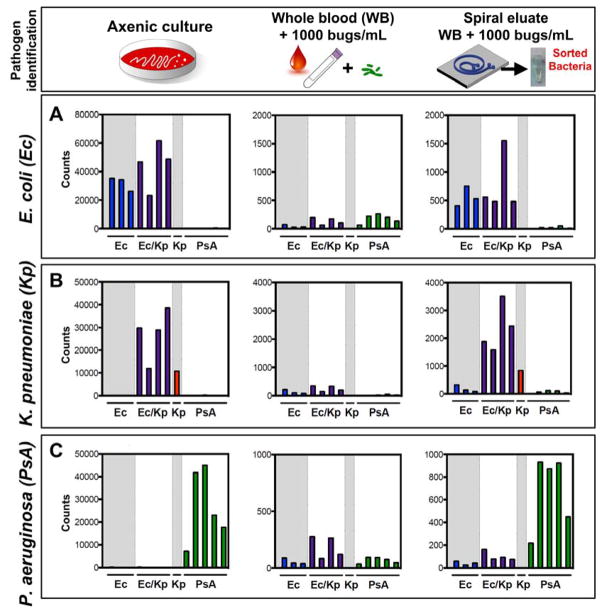
Figure 3.
Pathogen identification from blood after DFF by rRNA recognition. (A) E. coli, (B) K. pneumoniae, or (C) P. aeruginosa were either grown to mid-log phase in axenic culture and diluted 1:1000 (left panel); inoculated at 1000 cfu/mL in whole blood (middle panel); or inoculated at 1000 cfu/mL in whole blood, processed by DFF, and concentrated (right panel) before being detected by the NanoString assay using a probeset directed against rRNA from these three organisms (see Methods for details). On each graph, bars 1–3 represent E. coli-specific probes (Ec = blue), bars 4–7 represent probes that recognize either E. coli or K. pneumoniae but not P. aeruginosa (Ec/Kp = purple), bar 8 represents a K. pneumoniae-specific probe (Kp = red), and bars 9–13 represent P. aeruginosa-specific probes (PsA = green). Organism burden was enumerated by plating for CFU enumeration.