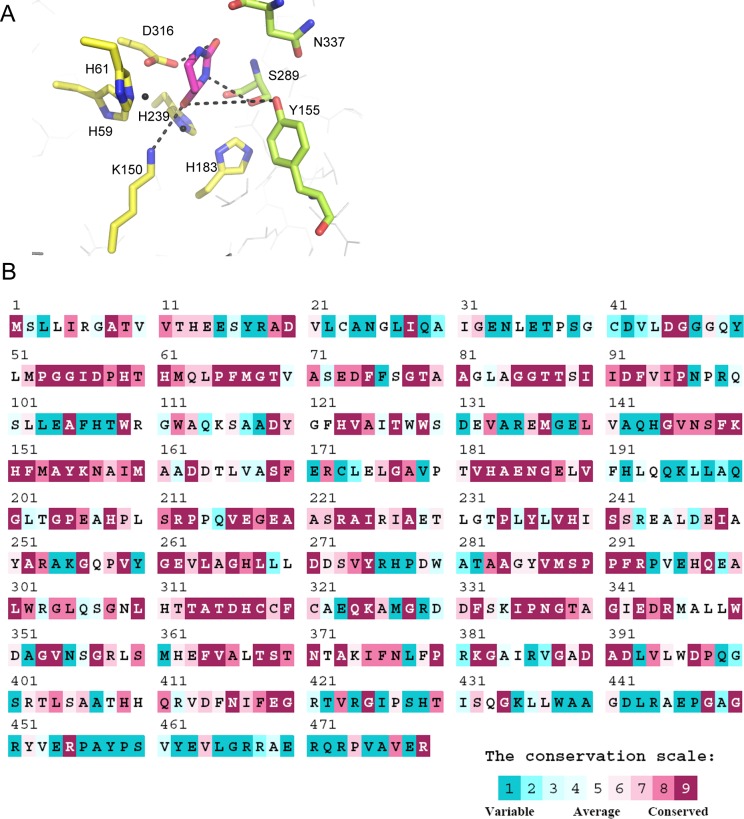
Fig 4. The active site of dihydropyrimidinase.
(A) The active site of P. aeruginosa dihydropyrimidinase. According to the crystal structure of Saccharomyces kluyveri (PDB entry: 2FVK), residues H59, H61, K150, H183, H239, and D316 of P. aeruginosa dihydropyrimidinase shown in yellow were crucial for the assembly of the binuclear metal center within the active site; meanwhile, residues Y155, S289, and N337 shown in limon were crucial for substrate binding. The model was directly constructed by superimposing the modeled structure of P. aeruginosa dihydropyrimidinase with the crystal structure of S. kluyveri dihydropyrimidinase-dihydrouracil complex. Dihydrouracil generated from the complex is shown in light magenta. (B) An alignment consensus of 497 sequenced dihydropyrimidinase homologs by ConSurf reveals the degree of variability at each position along the primary sequence. Note that the positions involved in the assembly of the binuclear metal center within the active site and the substrate binding of P. aeruginosa dihydropyrimidinase are well conserved.