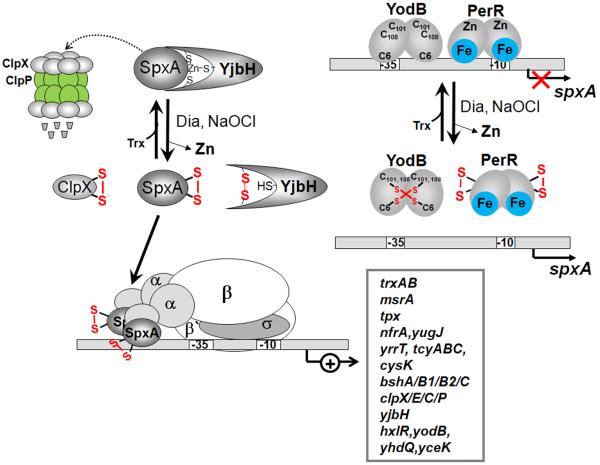
Figure 8. Post-translational and transcriptional control of SpxA by disulfide stress in B. subtilis.
Under non-stressed conditions, SpxA is unstable and targeted by the YjbH adaptor protein to the ClpXP machinery for proteolytic degradation. The stability of SpxA is increased by diamide due to oxidation of SpxA, YjbH and ClpX that prevents SpxA degradation. SpxA is oxidized in its redox-active CXXC motif and binds to the αCTD of RNAP resulting in transcriptional activation of the SpxA disulfide stress regulon. Transcription of spxA is regulated by the repressors YodB and PerR that are oxidized and inactivated under diamide stress leading to increased spxA transcription. SpxA controls a large regulon of genes encoding thioredoxin/thioredoxin reductase (trxAB), thiol peroxidase (tpx), FMN-dependent oxidoreductases (nfrA, yugJ), methionine sulfoxide reductase (msrA), cysteine biosynthesis enzymes (yrrT-operon, cysK, tcyABC), bacillithiol biosynthesis enzymes (bshA, bshB1, bshB2 and bshC), the protein quality control Clp machinery (clpX, clpE, clpC, clpP, yjbH) and several thiol-based redox regulators (hxlR, yodB, yhdQ, yceK). Examples for SpxA regulon genes and their functions are listed in Table 5.