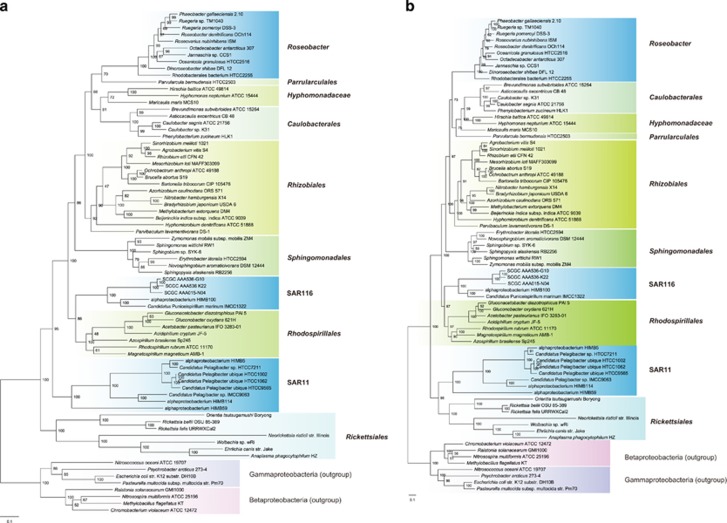
Figure 2.
Maximum likelihood phylogeny of Alphaproteobacteria using the RAxML v7.3.0 software. (a) Tree based on a concatenation of the 28 composition-homogeneous proteins, in which 19 are ribosomal proteins. (b) Tree based on a concatenation of the 24 composition-heterogeneous ribosomal proteins. A data partition model was employed to allow subsets of component proteins to evolve independently in amino acid replacement processes, which was determined using the PartitionFinder software. Values at the nodes show the number of times the clade defined by that node appeared in the 100 bootstrapped datasets. Trees are rooted using species from Betaproteobacteria and Gammaproteobacteria.