Figure 3.
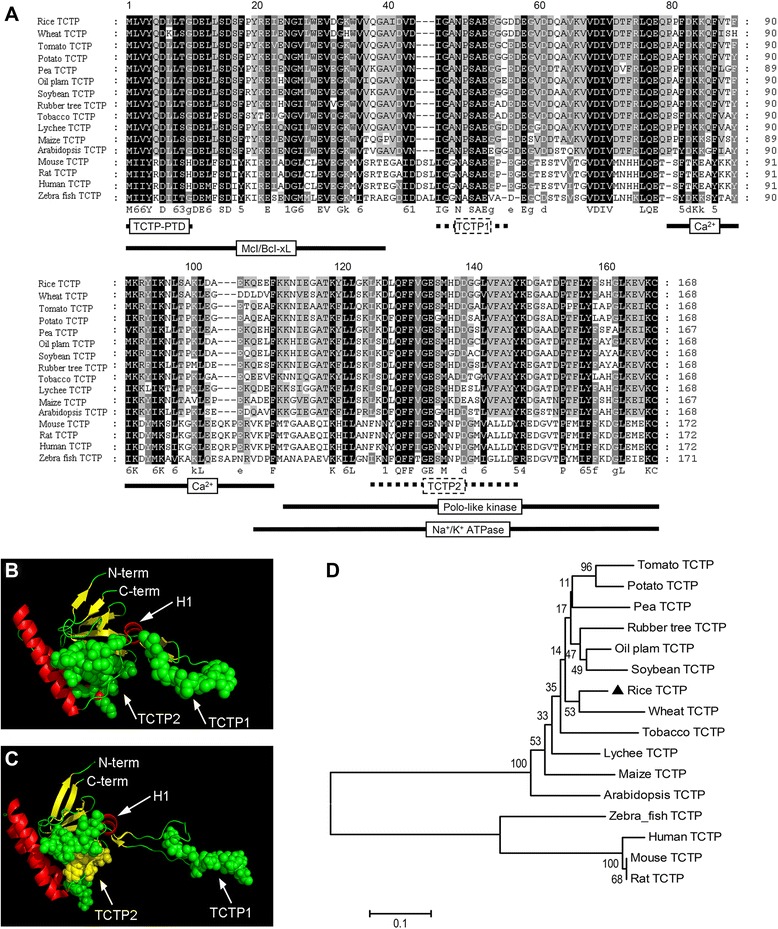
Sequence comparison, structural modeling and phylogenetic analysis of TCTP. (A) The amino acid sequence alignment was performed using the ClustalW2 software on representative plant and nonplant sequences. Positions with strictly conserved amino acids are highlighted in black, conserved substitutions in dark gray, and blocks of similar residues in light gray. Domains identified for nonplant TCTPs (Mcl/Bcl-xL interaction [42], Polo kinase interaction [43], Na+/K+ ATPase interaction [44], Ca2+ binding [45], TCTP self-interaction [46] and protein transduction domain [47]) are indicated by black lines and the TCTP signature by a dotted line. (B) The putative 3D structure of the OsTCTP. The structure of the OsTCTP protein was modeled using the known structure of the human TCTP (PDB ID 2HR9, http://www.rcsb.org) as a template on the Swiss-Model server (http://swissmodel.expasy.org [73]). The model obtained for the rice protein (b) shows high similarity when compared with the structure of the human protein (C). The conserved domains TCTP1 and TCTP2 of TCTP proteins are shown as calottes. The conserved helix (H1) is marked by an arrow. Helices, β-sheets, and coil regions of the two structures are represented in red, yellow, and green, respectively. (D) Phylogenetic analysis of OsTCTP and its 15 close homologs. The numbers under the branches refer to the bootstrap value of the neighbor-joining phylogenetic tree. The length of the branches is proportional to the amino acid variation rates. The scale bar indicates the number of amino acid substitutions per site.
