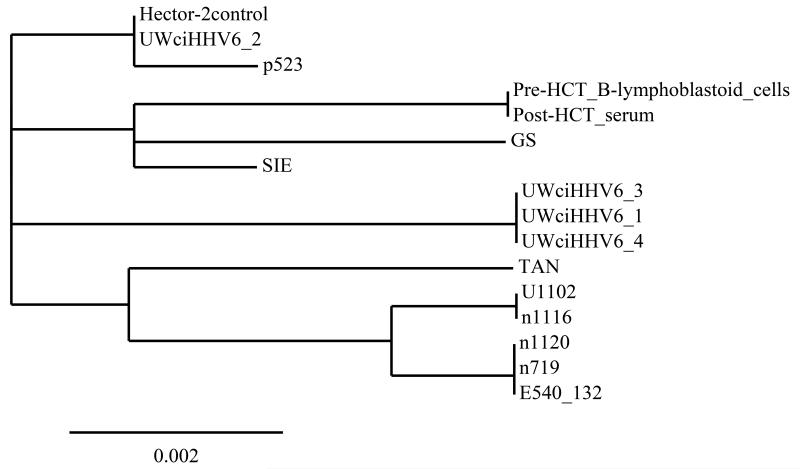
Figure 2.
Phylogenetic analysis of HHV-6A gB gene sequences from pre and post-HCT samples of the described patient with ciHHV-6A compared to ciHHV-6A from 4 patients at our center, the Hector-2 cell line (Bioworld Consulting Laboratories), and 9 published sequences8.
This figure demonstrates that the sequence of an ~900 base pair region of the HHV-6A gB gene in the discussed patient’s pre-HCT Beta-lymphoblastoid cell line and post-HCT (D+39) serum sample were identical and diverged from 14 other available sequences.
Bar = number of nucleotide changes per 100 sites.
The gB gene sequences of 9 HHV-6A primary or laboratory-adapted isolates were obtained from Dr Henri Agut (Paris, France), and the geographical origins were as follows: GS, United States; U1102, Uganda; SIE, Côte d’Ivoire; TAN, Congo; and 1120, E540_132, 1116, 719 and, p523, France. An additional 4 isolates were obtained from patients at the University of Washington with chromosomally integrated HHV-6: UWciHHV6_1, UWciHHV6_2, UWciHHV6_3, UWciHHV6_4.
The tree was constructed from the comparison of gB gene nucleotide sequences using the free Phylogeny.fr webservice, available at http://www.phylogeny.fr/version2_cgi/index.cgi.