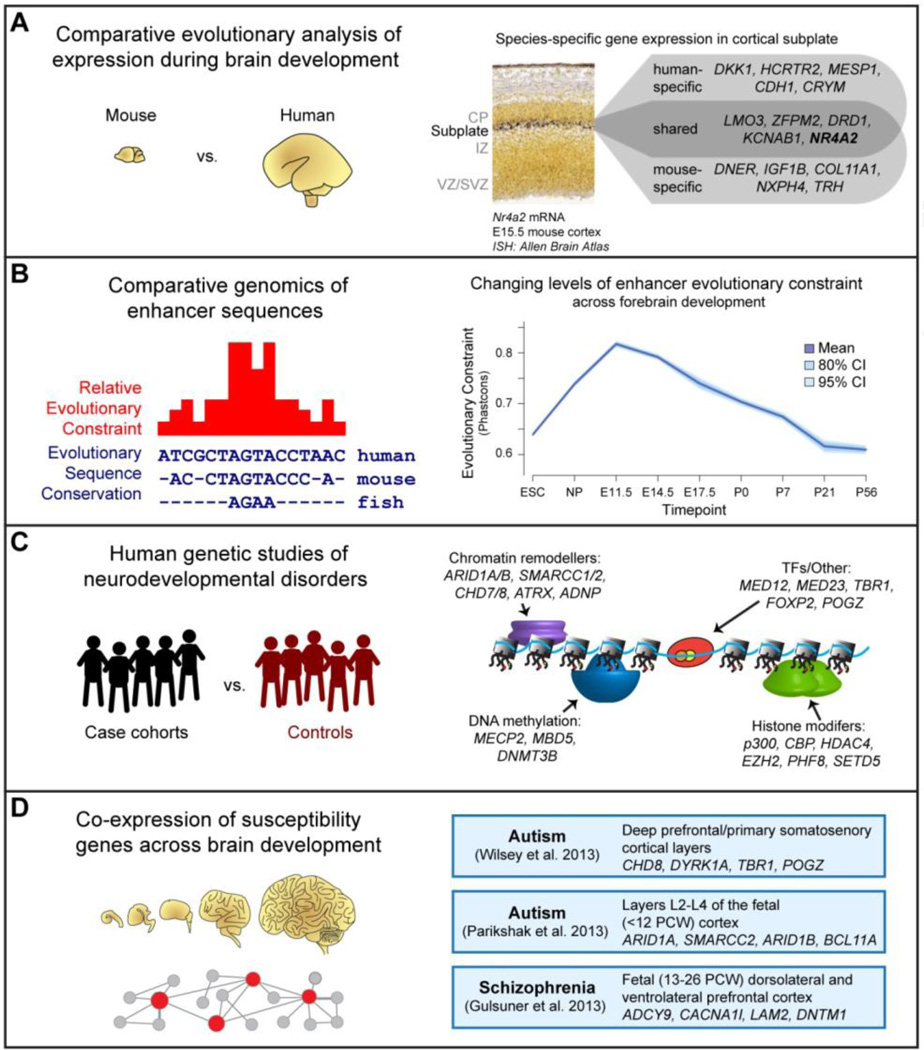
Figure 5. Insights and Challenges in Deciphering the Regulatory Architecture of Forebrain Development.
A) Comparative functional genomic studies, such as large-scale studies of gene expression patterns in the developing brain by RNA in situ hybridization, provide insight into general and human-specific aspects of molecular pathways involved in brain development. B) Comparative genomic studies reveal a deeply conserved regulatory framework associated with brain development, but can also identify specific changes in regulatory sequences that underlie structural and functional innovations in the brain observed in vertebrate evolution. Remarkably, regulatory sequences active during early stages of brain development (mid-gestation in mouse) tend to be under higher evolutionary constraint than those active later in development and in the adult brain. C) Genome-wide association, exome sequencing, copy number variation, and whole-genome sequencing studies of patient cohorts are powerful tools for identifying genes and non-coding sequences associated with neurodevelopmental disorders. These studies have revealed a major role for proteins involved in chromatin remodeling, DNA methylation processes, histone modification, and other transcriptional regulatory pathways and processes, with individual genes reported in human genetic studies offered as examples. D) Systems-level analysis integrating expression, genetic, epigenomic and functional data has the potential to elucidate genetic and functional networks required for normal brain development and function, which are thought to be disrupted in neurodevelopmental and neuropsychiatric disorders.