Abstract
Poly(β-amino ester)s (PBAEs) represent an important class of cationic gene delivery materials which, however, suffer from uncontrolled DNA release due in part to the slow degradation of their polyester backbone. Additionally, PBAEs with high molecular weight (MW) also show considerable toxicities. In this study, we designed and developed PBAEs with trigger-responsive domains built in polymer backbones that can be rapidly cleaved upon external UV light triggering to promote intracellular DNA release as well as reduce material toxicity. Photo-responsive PBAEs were prepared via polyaddition of (2-nitro-1,3-phenylene)bis(methylene) diacrylate and a bisfunctional amine. The nitrobenzene moiety was placed in each repeating unit of the PBAE to allow fast response to external UV irradiation, and thus the ester linkers were cleaved and the polymers were degraded within several minutes upon UV irradiation. Cationic PBAEs with high MWs were able to mediate effective intracellular gene delivery, while upon UV irradiation post-transfection, enhanced DNA unpackaging and reduced material toxicity were observed, which collectively contributed to greatly improved transfection efficiencies in various mammalian cell types tested. This strategy allows precise manipulation of material toxicity and gene release profiles of PBAEs, and thus provides an effective design approach to address critical issues in non-viral gene delivery.
Keywords: non-viral gene delivery, trigger responsiveness, poly(β-amino ester), degradable polymer, intracellular DNA release, cytotoxicity
1. Introduction
Gene therapy holds great promise for the treatment of congenital or acquired diseases by delivering generic materials into target cells to promote or rectify specific cellular functions [1–5]. Up- or down-regulation of specific gene targets have also been used to direct stem cell differentiation, promote tissue repairing, and reprogram somatic cells [6–8]. Gene delivery has been largely achieved with the use of viral vectors which are known for their high gene delivery efficiencies. However, the viral approach is often associated with significant immunogenicity, insertional mutagenesis, and oncogenicity, which presents serious concerns for its clinical applications [9, 10]. Non-viral gene delivery, known for its low immunogenicity and oncogenicity, has been intensively studied in the past two decades as a safer alternative to viral gene delivery [3, 5, 11–13]. Cationic polymers (or polycations), capable of condensing anionic nucleic acids to facilitate their intracellular delivery, are one of the most important classes of non-viral vectors. Polycations with higher molecular weight (MW) usually demonstrate stronger condensation capacity toward nucleic acids and mediate more efficient gene delivery than their lower MW analogues [14, 15]. However, they also show appreciable cytotoxicities related to their high MWs and cationic charge densities [16, 17]. Additionally, the excessive binding affinity of the high MW cationic polymer toward nucleic acids would also restrict the intracellular gene release, a critical roadblock toward effective gene transfection [18].
Poly(β-amino ester) (PBAE) is one of the few polycations that have been very intensively studied in non-viral gene delivery in the past decade. It is a class of biodegradable polymers designed by Langer and co-workers that can be easily obtained in large scale via elegantly simple yet extremely versatile polyaddition chemistry, which can be readily adapted to high-throughput processes to make a large library of structurally diverse materials [19–25]. Optimal structures have also been identified that lead to efficient gene delivery to a variety of mammalian cells in vitro and to eyes and tumors in vivo [26, 27]. Because PBAEs are comprised of degradable ester linkers on the backbone, they are hydrolysable and therefore polymer degradation may trigger intracellular release of the gene cargos. However, the hydrolysis of the polyester backbone occurs on the time scale of several hours to a few days, and is largely affected by the polymer structure as well as the cellular condition [22, 28, 29]. As such, it is unlikely to control over when and where gene release will occur. For instance, excessively rapid polymer degradation may lead to undesired pre-release of the gene cargos in the extracellular compartment to hamper their cellular internalization, while slow degradation profile up to several days may retard the intracellular release to impair the transfection efficiency. Given the drawback of such un-controlled release mechanism, it is of great interest to precisely control the polymer degradation to allow “on-demand” cargo release at a specific intracellular process (e.g. in the cytoplasm).
To realize this goal, we herein developed trigger-responsive PBAEs containing light-responsive 2-nitrobenzene moieties in the polymer backbone. We hypothesized that these trigger-responsive PBAEs with high MWs and cationic charge densities can efficiently condense and deliver genes intracellularly; while upon external triggering at the post-transfection stage, the polymeric backbone can be rapidly degraded into small fragments such that intracellular gene release can be facilitated and material toxicity associated with high MW and charge density can be reduced. To this end, a small library of trigger-responsive PBAEs was synthesized via condensation of (2-nitro-1,3-phenylene)bis(methylene) diacrylate (NPBMDA) and various bisfunctional amines. The best-performing material with optimal structure was first identified toward gene transfection, and it was further subjected to evaluation on the trigger-responsive gene delivery properties, such as polymer degradation profiles, DNA release, intracellular kinetics, gene transfection efficiency, and cytotoxicity.
2. Materials and methods
2.1. Materials and cell lines
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received unless otherwise specified. Anhydrous tetrahydrofuran (THF), dichloromethane (DCM), hexane, and dimethylformamide (DMF) were dried by a column packed with 4Å molecular sieves before use. Pierce BCA assay kit was purchased from ThermoFisher Scientific (Rockford, IL, USA). Plasmid DNA (pDNA) encoding enhanced green fluorescence protein (EGFP) (pEGFP) was purchased from Elim Biopharm (Hayward, CA, USA). Lipofectamine™ 2000 (LPF), 3-(4,5-dimethylthiahiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT), and YOYO-1 were purchased from Invitrogen (Carlsbad, CA, USA).
HeLa (human cervix adenocarcinoma cells), COS-7 (African green monkey kidney cells), and 3T3-L1 (mouse embryonic fibroblast) were purchased from the American Type Culture Collection (Rockville, MD, USA) and were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (for HeLa and COS-7 cells) or 10% bovine calf serum (for 3T3-L1 cells).
2.2. Synthesis and characterization of monomers
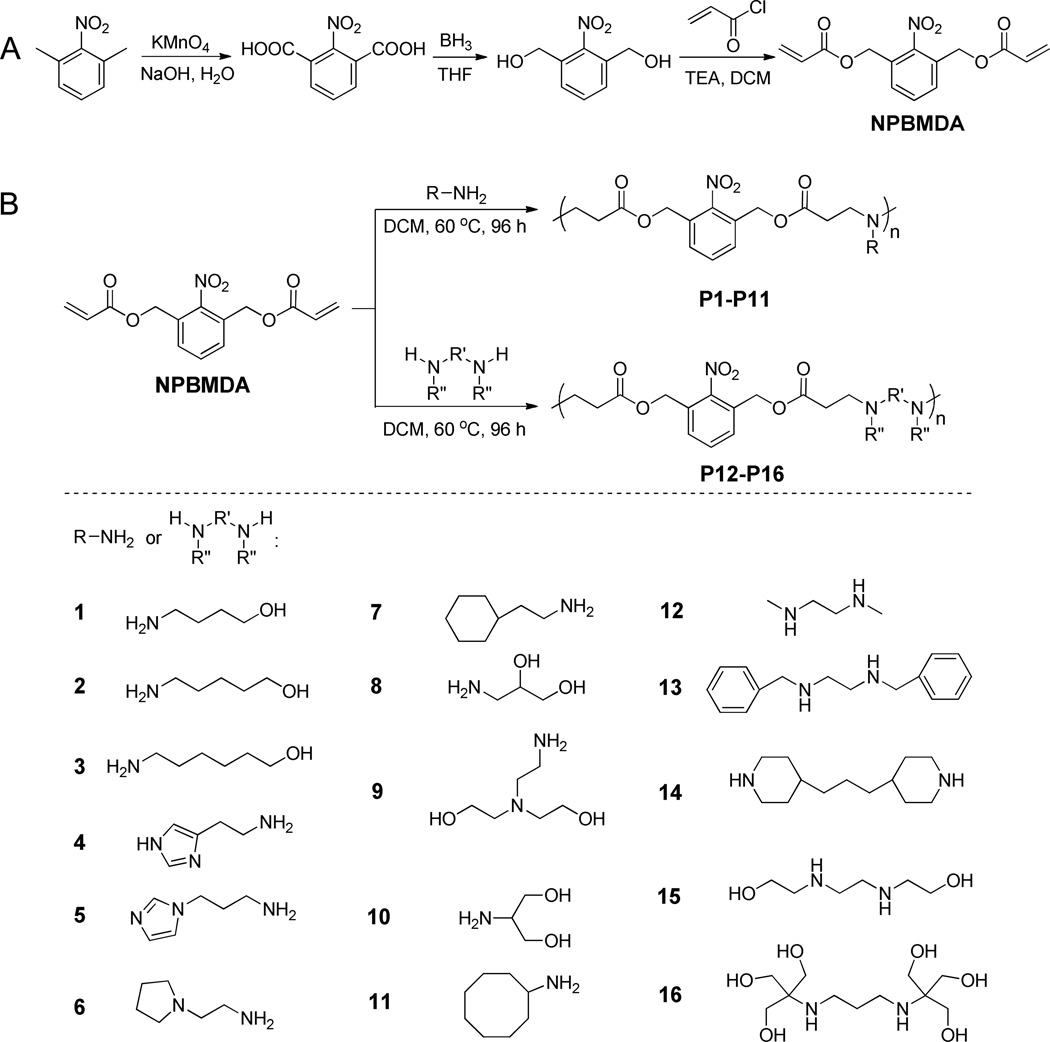
2-Nitro-1,3-benzenedimethanol was synthesized as illustrated in Scheme 1A. 1,3-Dimethyl-2-nitrobenzene (15.0 g, 0.10 mol) was added to a stirred NaOH basic solution (0.2 M, 800 mL) at 95 °C. KMnO4 (66 g, 0.418 mol) was then slowly added, and the resulting mixture was refluxed for 24 h. The mixture was then filtered after cooling to room temperature and the filtrate was acidified with HCl to pH 1 to obtain the product 2-nitro-1,3-benzenedicarboxylic acid as white solid (11.0 g, yield 52%). 1H NMR (DMSO-d6): δ 8.17 (m, 2H, ArH), 7.79 (m, 1H, ArH).
Scheme 1.
Synthesis of (2-nitro-1,3-phenylene)bis(methylene) diacrylate (A) and a library of photo-responsive PBAEs (P1–P16) with various side charged groups.
2-Nitro-1,3-benzenedicarboxylic acid (16.0 g, 76 mmol) was dissolved in anhydrous THF (100 mL) and cooled to 4 °C in an ice bath. Borane (1.0 M in THF complex solution, 400 mL) was slowly added by syringe over 1 h under N2, and the reaction mixture was warmed to room temperature and stirred for another 48 h. Methanol (40 mL) was then added dropwise to the reaction mixture. The mixture was filtered and dried under vacuum. The residue was re-dissolved in EtOAc and washed with saturated NaCl solution (4 × 100 mL). The organic layer was dried with anhydrous MgSO4 for 12 h before the solvent was removed under vacuum. The crude product was obtained as yellow solid, which was further purified by silica gel chromatography (hexane:EtOAc as eluent, 1:1, v/v) to obtain 2-nitro-1,3-benzenedimethanol (11.0 g, yield 80%). 1H NMR (DMSO-d6): δ 7.68 (m, 3H, ArH), 5.56 (t, 2H, -OH), 4.70 (d, 4H, -CH2OH).
To synthesize (2-nitro-1,3-phenylene)bis(methylene) diacrylate (NPBMDA), the designated light-responsive monomer, triethylamine (100 mmol), was added dropwise into a solution of 2-nitro-1,3-benzenedimethanol (7.3 g, 40 mmol) in anhydrous DCM (50 mL) over 1 h under N2. Acryloyl chloride was slowly added into the reaction mixture by syringe. The mixture was stirred for 18 h at room temperature and filtered. The filtrate was dried under vacuum and the residue was then re-dissolved in EtOAc. The resulting solution was washed with saturated NaCl solution (3 × 100 mL). The organic layer was dried with anhydrous MgSO4 overnight before the solvent was removed under vacuum. The crude product was obtained as yellow solid which was further purified by silica gel chromatography (hexane:EtOAc as eluent, 1:1, v/v) to obtain NPBMDA as white crystal (8.1 g, yield 70%). 1H NMR (DMSO-d6): δ 7.65 (m, 3H, ArH), 6.28 (d, 2H, -CH=CH2), 6.12 (dd, 2H, -CH=CH2), 5.95 (d, 2H, -CH=CH2), 5.23 (s, 4H, ArCH2O-).
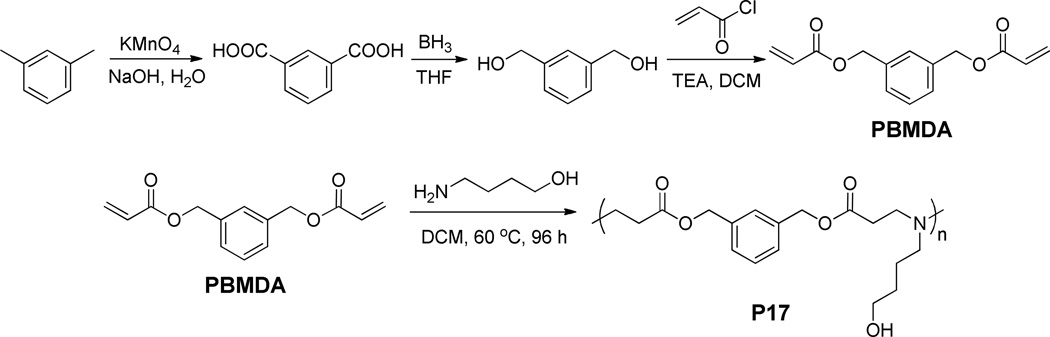
(1,3-Phenylene)bis(methylene) diacrylate (PBMDA) as the control, non-responsive monomer was synthesized by following the same method as described above with m-tolunitrile instead of 1,3-dimethyl-2-nitrobenzene as the substrate (Scheme 2). The final product PBMDA was obtained as colorless viscous liquid (yield 56%). 1H NMR (DMSO-d6): δ 7.36 (m, 3H, ArH), 6.34 (d, 2H, -CH=CH2), 6.20 (dd, 2H, -CH=CH2), 5.94 (d, 2H, -CH=CH2), 5.16 (s, 4H, ArCH2O-).
Scheme 2.
Synthesis of (1,3-phenylene)bis(methylene) diacrylate and the corresponding non-responsive PBAE (P17).
2.3. Synthesis and characterization of PBAEs
Light-responsive PBAEs were synthesized from NPBMDA and various amine-containing compounds via Michael addition reaction (Scheme 1B and Supplementary Table S1). Briefly, NPBMDA (0.4 mmol) and each individual amine-containing molecule (amine/diacrylate molar ratio = 1.05~1.3) were dissolved in DCM (2 mL) and the reaction was allowed to proceed at 60 °C for 4 days. After removal of the solvent under vacuum, the polymers were purified by precipitation with methanol and isolated as yellow solid or viscous liquid (yield 31~52%). DMSO was selected as the solvent for reactions using 2-(1H-imidazol-4-yl)ethanamine and 3-(1H-imidazol-1-yl)propan-1-amine that cannot dissolve in DCM, and the obtained polymers were purified by precipitation with ether. The MWs of the obtained polymers were measured by GPC. The obtained polymers were named as Pn-m, where n (1~16) is the specific amine type as shown in Scheme 1B and m is the MW (Da).
To enable direct comparison on the light-responsiveness of PBAEs, P17, a non-responsive analogue of P1, was synthesized from the non-responsive monomer PBMDA and 4-amino-1-butanol at the starting feed ratio of 1.08 using the same method described above (Scheme 2).
2.4. UV-triggered polymer degradation
Polymers were dissolved in DMF at 10 mg/mL, placed in a quartz vial, and irradiated with UV light (λ = 365 nm, 20 mW/cm2) for different periods of time. The UV-Vis spectra were recorded to monitor the photo-triggered polymer degradation and the generation of nitrosobenzene derivatives. The MWs of the UV-irradiated polymers were determined by GPC to confirm the polymer degradation.
2.5. Preparation and characterization of polyplexes
Polymers were dissolved in DMSO at 100 mg/mL and diluted with sodium acetate buffer (25 mM, pH 5.2) to 1 mg/mL. The polymer solution was then added to DNA (0.2 mg/mL) at various pre-selected weight ratios, vortexed for 30 s, and incubated at room temperature for 20 min to allow polyplex formation. The polyplexes were subjected to electrophoresis in 1% agarose gel at 100 mV for 45 min to evaluate DNA condensation by the polymers in terms of DNA migration. To quantitatively monitor the DNA condensation level, the ethidium bromide (EB) exclusion assay was adopted [14]. Briefly, polyplexes were prepared and EB solution was added at the DNA/EB ratio of 10:1 (w/w). After incubation at room temperature for 1 h, the fluorescence intensity was monitored on a microplate reader (λex=510 nm, λem=590 nm). A pure EB solution and the DNA/EB solution without any polymer served as negative and positive controls, respectively. The DNA condensation efficiency (%) was defined as:
FEB, F, and F0 denote the fluorescence intensity of pure EB solution, DNA/EB solution with polymer, and DNA/EB solution without any polymer, respectively. The particle size and zeta potential of freshly prepared polyplexes were also evaluated by dynamic laser scanning (DLS) on a Malvern Zetasizer (Malvern Instruments Inc., Herrenberg, Germany).
2.6. UV-triggered polyplex dissociation and DNA release
Freshly prepared polyplexes (polymer/DNA weight ratio of 10) were irradiated by UV light (λ = 365 nm, 20 mW/cm2) for different periods of time and thereafter subjected to particle size analysis by DLS. The heparin replacement assay was adopted to evaluate the UV-triggered DNA release from the polyplexes [30]. Briefly, heparin was added to the non-irradiated and the UV-irradiated polyplexes (polymer/DNA weight ratio of 10) solution at various final concentrations and the solutions were incubated at 37 °C for 1 h. The DNA condensation level was quantified using the EB exclusion assay as described above.
2.7. In vitro transfection
Cells were seeded on 24-well plates at 5 × 104 cells/well and cultured in serum-containing media for 24 h before reaching confluence. The culture medium was changed to Opti-MEM (500 µL/well) into which polyplexes were added at 0.5 µg DNA/well. After incubation at 37 °C for 4 h, the medium was replaced by DMEM containing 10% FBS (500 µL/well), irradiated with UV light (λ=365 nm, 20 mW/cm2) for 2 min, and further incubated for 48 h before assessment of EGFP expression by flow cytometry. The transfection efficiency was expressed as the percentage of EGFP-positive cells (%). The EGFP expression was also observed by fluorescence microscopy.
2.8. Intracellular kinetics
To allow quantification of the cellular uptake level, DNA (1 mg/mL) was labeled with YOYO-1 (20 µM) at one dye molecule per 50 bp DNA [31]. The resultant YOYO-1-DNA was allowed to form polyplexes at various N/P ratios as described above. Cells were seeded on 24-well plates at 5 × 104 cells/well, and cultured for 24 h to reach confluence. The medium was replaced by Opti-MEM and polyplexes were added at 0.5 µg YOYO-1-DNA/well. After incubation at 37 °C for 1, 2, and 4 h, cells were washed with PBS three times before treated with the RIPA lysis buffer (500 µL). YOYO-1-DNA content in the lysate was quantified by spectrofluorimetry (λex=485 nm, λem=530 nm) and protein content was measured using the BCA kit. Uptake level was expressed as ng YOYO-1-DNA associated with 1 mg cellular protein. The cellular internalization and distribution of YOYO-1-DNA was also observed by confocal laser scanning microscopy (CLSM). Briefly, HeLa cells were incubated with polymer/YOYO-1-DNA polyplexes in Opti-MEM at 0.5 µg DNA/well (6-well plate) for 4 h. Cells were then washed three times with PBS, fixed with paraformaldehyde (4%), stained with DAPI (2 µg/mL, for nuclei) and Lysotracker Red® (200nM, for endosome/lysosome), and visualized by CLSM (LSM 700, Zeiss, Germany).
To elucidate the mechanisms underlying the cellular internalization of polyplexes, the uptake study was performed at 4 °C or in the presence of various endocytic inhibitors within a period of 2 h. Briefly, cells were pre-incubated with one of the selected endocytic inhibitors (chlorpromazine (10 µg/mL), genistein (200 µg/mL), methyl-β-cyclodextrin (mβCD, 50 µM), dynasore (80 µM), and wortmannin (50 nM)) for 30 min prior to the addition of polyplexes and throughout the 2-h uptake experiment at 37 °C. Results were expressed as percentage uptake level of control cells that were incubated with complexes at 37 °C for 2 h in the absence of endocytic inhibitors.
2.9. Cytotoxicity
Cells were seeded on 96-well plates at 1 × 104 cells/well and cultured in serum-containing media for 24 h. The medium was replaced with Opti-MEM (100 µL/well) into which polyplexes at various polymer/DNA weight ratios were added at 1 µg DNA/mL. After incubation at 37°C for 4 h, the medium was changed to serum-containing DMEM. The cells were irradiated with UV light (λ = 365 nm, 20 mW/cm2) for 3, 5, or 10 min, cultured for additional 48 h, and evaluated for viability using the MTT assay. Cells without UV irradiation served as a control. Results were represented as percentage viability of cells that did not receive polyplex treatment or UV irradiation.
2.10. Statistical analysis
Statistical analysis was performed using Student’s t-test. The differences between test and control groups were assessed to be significant at *p < 0.05 and very significant at **p < 0.01.
3. Results and discussion
3.1. Synthesis of trigger-responsive PBAEs
For gene delivery purposes, it is ideal that degradation of polycations proceed rapidly upon internal/external triggering so that DNA release can be precisely localized at the specific intracellular phase and subcellular compartment. To this end, we designed PBAEs that contain a light-responsive moiety in each repeating unit of the polymer backbone. Because each repeating unit has a cleavable site, the sensitivity towards light irradiation is high and the polymer can be rapidly degraded into small pieces to trigger “on-demand” DNA release. To realize such design strategy, NPBMDA, a light-responsive monomer, was first synthesized from 1,3-dimethyl-2-nitrobenzene (Scheme 1A) and its molecular structure was confirmed by 1H NMR (Supplementary Fig. S1). PBMDA, the non-responsive analogue of NPBMDA, was synthesized from m-tolunitrile in a similar manner (Scheme 2, Supplementary Fig. S2) and used as the monomer to synthesize the control PBAE.
The macromolecular structures of PBAEs, such as MW, end group, and amine charge group, all should have notable effect on their gene delivery capabilities [32, 33]. Thus, we first attempted to identify a suitable structure with sufficiently high gene transfection efficiency by synthesizing and screening a small library of light-responsive PBAEs via Michael-type polyaddition reactions of NPBMDA and various structure- and property- (e.g., hydrophobicity/hydrophilicity) diverse amines (Scheme 1B). Although highest MW should be achieved at the amine/diacrylate molar ratio of 1, previous studies have demonstrated that amine-terminated PBAEs possess higher transfection efficiencies [34]. Therefore, we fixed the amine/diacrylate molar ratio at 1.05~1.3 in this study so that PBAEs with amine end groups can be synthesized. By changing the amine/diacrylate ratios, we synthesized P1-P16 with MWs ranging from 3 to 14 kDa (Scheme 1, Supplementary Table S1). P1–P16 are all yellowish solid or liquid, and are well soluble in aqueous solution. P17–11500, a non-responsive analogue of P1 with similar structure, charge density, and MW, was prepared from the non-responsive monomer PBMDA and 4-amino-1-butanol (Scheme 2).
3.2. Screening PBAE gene delivery efficiency
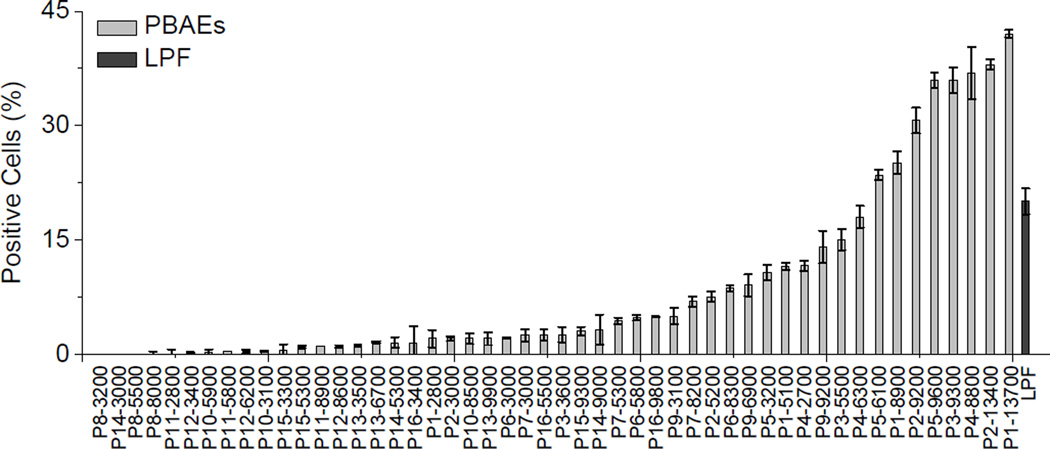
We first studied the in vitro gene transfection efficiencies of P1–P16 in HeLa cells, in attempts to identify a suitable polymer for further study on trigger-induced DNA unpackaging and toxicity reduction. As shown in Fig. 1, PBAEs bearing primary hydroxyl side groups and hydrophobic alkyl chains (P1–P3) exhibited relatively high transfection efficiencies. Polycations containing imidazole moiety have been reported to have strong buffering capacities and may facilitate endosomal escape via the “proton sponge” effect [35]. PBAEs containing imidazole side group (P4 and P5) were thus synthesized and found to show transfection efficiencies comparable to P1–3. In general, PBAEs with higher MWs show higher gene transfection efficiencies, presumably due to formation of more stable complex and more efficient internalization with the use of polycations with high MW and cationic charge densities [33, 36, 37]. Up to 42% of HeLa cells were transfected with the use of P1–13700 while less than 5% of cells were transfected by P1–2800.
Fig. 1.
Transfection efficiencies of PBAEs with various side charged groups in HeLa cells. Results were represented as percentage (%) of EGFP-positive cells.
3.3. UV-triggered degradation of PBAEs
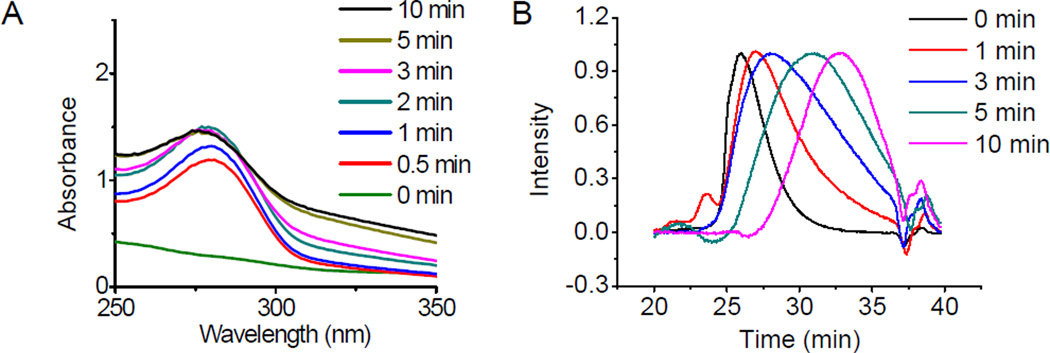
P1–13700, the PBAE showing highest gene transfection efficiency in all materials tested, was selected for further investigation. We first studied light-triggered polymer degradation using both UV-Vis spectrometry and GPC. When a DMF solution of P1–13700 was irradiated with UV light (λ = 365 nm, 20 mW/cm2) for as short as 30 s, a significantly increase of the absorbance at 280 nm (OD280) was observed (Fig. 2A), which indicated the generation of the nitrosobenzene group due to cleavage of the photo-labile ester bond (Scheme 3). At the increase of the irradiation time, a further increase of the OD280 was noted. P17–11500, the control PBAE, showed unappreciable alteration of the OD280 in the UV-Vis spectra upon UV irradiation (Supplementary Fig. S4), which indicated that UV irradiation cannot cleave other non-UV active segments of the polymer. The degradation of P1–13700 was further confirmed by GPC analysis. As illustrated in Fig. 2B and Supplementary Fig. S5, the MWs of P1–13700 were dramatically decreased upon UV treatment and a 36% reduction of the MW was noted post 10-min irradiation (Supplementary Fig. S5). In comparison, the UV non-responsive P17–11500 cannot be degraded by UV irradiation and thus its MW remained unchanged upon UV irradiation (data now shown).
Fig. 2.
Light-triggered degradation of PBAEs. UV-Vis spectra (A) and GPC curves (B) of P1–13700 after UV irradiation (λ = 365 nm, 20 mW/cm2) for various time.
Scheme 3.
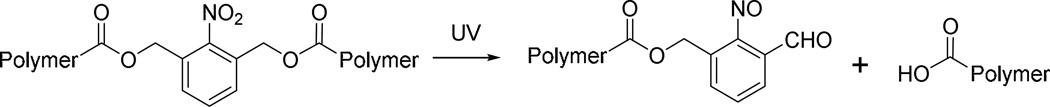
Proposed degradation mechanism of PBAEs in response to UV irradiation.
3.4. Polyplex formation and UV-triggered DNA release
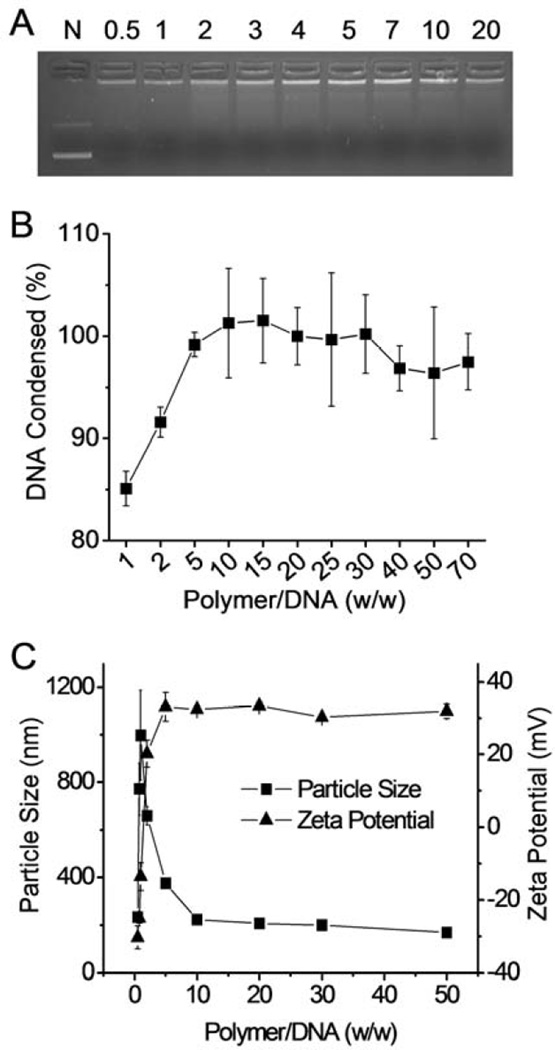
The capacity of the cationic PBAE to condense DNA was first evaluated using the gel retardation assay with P1–13700 as the representative polymer. As shown in Fig. 3A, DNA migration in the 1% agarose gel was completely restricted to the loading well at the polymer/DNA weight ratio higher than 2, indicating complete condensation of DNA by the positively charged PBAE. Such results were further verified by a quantitative EB exclusion assay, wherein more than 80% of the DNA was condensed at the polymer/DNA weight ratio of 1 and complete DNA condensation occurred at the polymer/DNA weight ratio higher than 10 (Fig. 3B). Consistently, DLS measurement revealed that stable polyplexes were formed at the polymer/DNA weight ratio higher than 10 with diameter of ~ 200 nm and positive surface charge of ~ 35 mV (Fig. 3C).
Fig. 3.
Characterization of PBAE/DNA polyplexes. DNA condensation by P1–13700 at different polymer/DNA weight ratios as evaluated by the gel retardation assay (A) and the EB exclusion assay (B). N represents naked DNA in (A). (C) Size and zeta potential of P1–13700/DNA polyplexes.
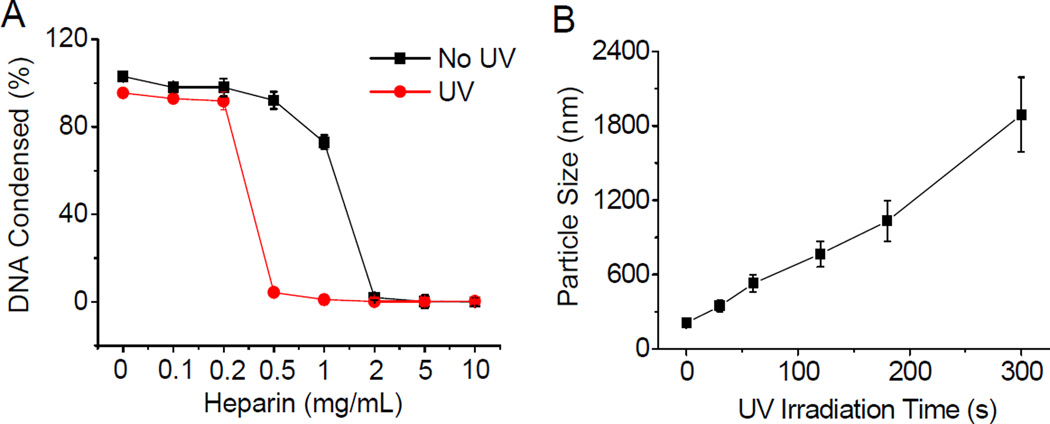
UV-induced DNA unpackaging was then monitored using the heparin replacement assay. As shown in Fig. 4A, DNA was completely released from UV-irradiated polyplexes (20 mW/cm2, 5 min) in the presence of 0.5 mg/mL heparin, while in comparison, higher heparin concentration of 2 mg/mL was required to fully dissociate DNA from the non-irradiated polyplexes. Such discrepancy clearly demonstrated that the DNA binding affinity of the UV-responsive P1–13700 was notably reduced after UV irradiation. As a control, the non-responsive P17–11500 showed unappreciable alteration in terms of heparin-induced DNA release profiles upon UV treatment (Supplementary Fig. S6), which verified that light-triggered polymer degradation contributed to the promoted DNA unpackaging. In accordance with the polymer degradation profiles, longer UV irradiation correlated to faster DNA release profiles in the absence of heparin within the observation period of 48 h (Supplementary Fig. S7), which further confirmed that higher degradation levels could accelerate the DNA release rate. In support of these quantitative results, promoted DNA migration in the agarose gel was also observed after 5-min UV irradiation of the P1–13700/DNA complexes but not the P17–11500/DNA complexes (Supplementary Fig. S8). In consistence with the UV-triggered DNA dissociation, particle size of the P1–13700/DNA polyplexes but not the P17–11500/DNA polyplexes was also markedly augmented upon UV treatment, which signified reduced binding affinity of PBAE toward DNA that served as the driving force for polyplex formation (Fig. 4B). These results collectively substantiated our design strategy to promote “on-demand” DNA release by cascading instantaneous polymer degradation using external triggers.
Fig. 4.
Light-triggered polyplex dissociation and DNA release. (A) DNA release from UV-irradiated and nonirradiated P1–13700/DNA polyplexes in the presence of heparin at various concentrations (n = 3). (B) Alteration of P1–13700/DNA polyplex diameter upon UV irradiation (λ = 365 nm, 20 mW/cm2) for different time periods as measured by DLS.
3.5. In vitro transfection
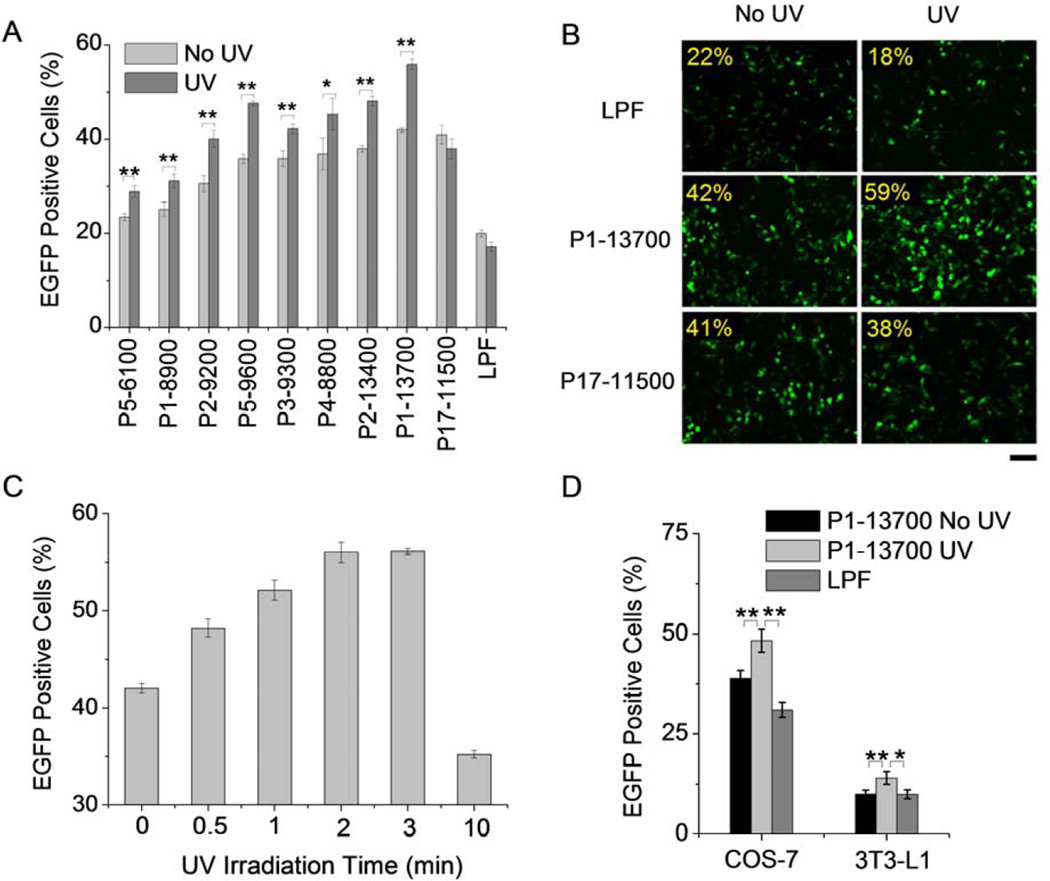
A couple of trigger-responsive PBAEs were firstly selected and subjected to transfection assessment in HeLa cells upon irradiation with UV, an efficient model light trigger. UV irradiation was performed at the post-transfection state after 4-h polyplex treatment, wherein effective cellular internalization of the polyplexes had been achieved. As shown in Fig. 5A, UV irradiation for 2 min post-transfection led to significant enhancement in the transfection efficiencies of all tested UV-responsive PBAEs, demonstrating that UV-triggered backbone cleavage potentiated gene transfection by facilitating the intracellular DNA release. UV-triggered P1–13700 exhibited the highest transfection efficiencies among all test materials, and in a direct comparison with LPF as the benchmark positive control, a 2–3 fold improvement was also noted. UV treatment did not notably alter the transfection efficiencies of LPF and P17–11500, which further demonstrated that light treatment itself did not improve gene expression. Such results were also confirmed by fluorescence images (Fig. 5B), where UV treatment augmented the GFP expression level of P1–13700 but not the controls, including P17–11500 and LPF. It was further noted that prolonged UV irradiation led to greatly increased transfection efficiency which plateaued at 2-min irradiation (Fig. 5C and Supplementary Fig. S9). It therefore indicated that 2-min irradiation was sufficient to provoke intracellular DNA release towards maximal transfection. Excessively long time irradiation may impair cell integrity, and thus the transfection efficiency decreased rather than increased. Upon identifying the light-promoted gene transfection profiles of the UV-responsive PBAEs, we were further motivated to demonstrate the generality of this approach in other mammalian cell types, including COS-7 and 3T3-L1 cells. Consistently, UV irradiation for 2 min induced significantly elevated transfection efficiencies of P1–13700/DNA polyplexes which outperformed LPF as the positive control (Fig. 5D). Considering the potential toxicity of UV light, we also monitored the cell viability upon UV irradiation. Cells receiving low intensity UV irradiation for a short period of time (20 mW/cm2, 2 min) exhibited uncompromised viability (96.8 ± 4.2%), indicating that UV irradiation in this proof-of-concept model system did not induce appreciable cytotoxicity that would otherwise jeopardize the analysis of trigger-responsive gene delivery capabilities.
Fig. 5.
Light-responsive transfection capabilities of PBAE/DNA polyplexes (weight ratio of 50). (A) A comparison of transfection efficiencies of PBAE/DNA polyplexes in HeLa cells in response to UV irradiation (λ = 365 nm, 20 mW/cm2) for 2 min. Results were represented as percentage (%) of EGFP-positive cells (n = 3). (B) Fluorescence images of transfected HeLa cells w/ and w/o UV irradiation. Bar represented 50 µm. (C) Effect of UV irradiation time on the transfection efficiencies of P1–13700/DNA polyplexes (n = 3). Laser power was fixed at 20 mW/cm2 (λ = 365 nm). (D) Enhanced transfection efficiencies of P1–13700/DNA polyplexes in COS-7 and 3T3-L1 cells in response to UV irradiation for 2 min (λ = 365 nm, 20 mW/cm2, n = 3).
3.6. Intracellular kinetics
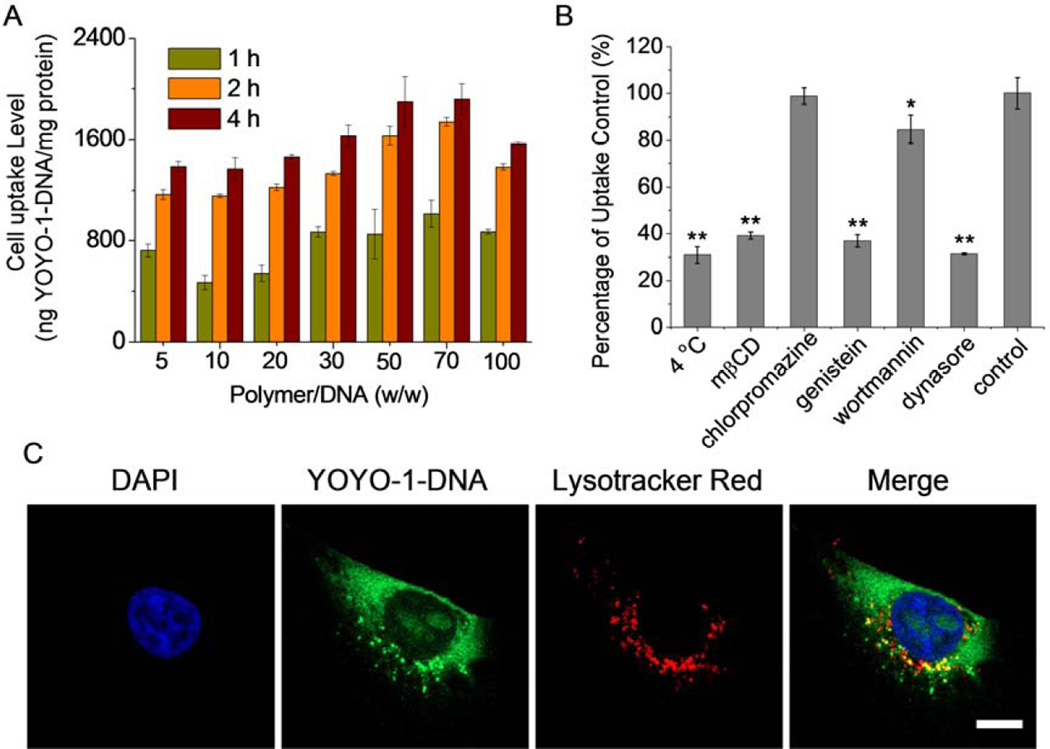
The gene transfection efficiencies of non-viral vectors are closely related to their intracellular kinetics and internalization pathways [38]. After demonstrating enhanced gene transfection upon UV treatment, we went on to study the cellular uptake and the internalization mechanisms of the polyplexes formed by P1–13700 and YOYO-1 labeled DNA (YOYO-1-DNA). As shown in Fig. 6A, P1–13700 remarkably promoted the internalization of DNA in HeLa cells, and the maximal uptake level was noted at the polymer/DNA weight ratio of 50~70. CLSM images further revealed that polyplexes were effectively taken up by HeLa cells, and the cellular uptake level was greatly enhanced with prolonged incubation time, as evidenced by the cytoplasmic distribution of green fluorescence (YOYO-1-DNA, Supplementary Fig. S10). Nuclear distribution was also noted post 4-h incubation, indicating that some released DNA was transported to the nuclei where gene transcription was triggered.
Fig. 6.
Intracellular kinetics of P1–13700/YOYO-1-DNA polyplexes (weight ratio of 50) in HeLa cells. (A) Uptake level of polyplexes following incubation at 37 °C for different time (n = 3). (B) Uptake level of polyplexes at 4 °C or in the presence of various en docytic inhibitors. Results were represented as percentage (%) of the uptake level at 37 °C and in the absence of inhibitors (n = 3). (C) CLSM images showing the cellular internalization and distribution of polyplexes in HeLa cells following incubation at 37 °C for 4 h. Cell nuclei were stained with DAPI and the endosomes/lysosomes were stained with Lysotracker Red. Bar represented 10 µm.
We then probed the internalization mechanism and pathway of the polyplexes by performing the cell uptake study at lower temperature (4 °C) or in the presence of various endocytic inhibitors. Energy-dependent endocytosis should be completely blocked at 4 °C; chlorpromazine inhibits clathrin-mediated endocytosis (CME) by triggering the dissociation of the clathrin lattice; genistein and mβCD prevent caveolae pathway by inhibiting tyrosine kinase and depleting cholesterol, respectively; dynasore inhibits both CME and caveolae by inhibiting dynamin; wortmannin inhibits macropinocytosis by suppressing phosphatidyl inositol-3-phosphate [38–40]. As shown in Fig. 6B, cell uptake was notably decreased by 70% at 4 °C, indicating that majority of the polyplexes were internalized via energy-dependent endocytosis, and the remaining were adsorbed onto cell surfaces via physical binding. The cell uptake level was also significantly reduced by genistein, mβCD, and dynasore but not chlorpromazine, indicating that polyplexes were internalized mainly via coavelae rather than CME. Wortmannin showed slight inhibitory effect, implying that macropinocytosis was also involved during endocytosis. Caveolar uptake is a non-acidic and non-digestive route of internalization, and cargos in the caveosomes can be directly transported to the Golgi and/or endoplasmic reticulum [38]. Macropinosome membrane is inherently leaky, which also allows direct escape of the entrapped cargos in the cytoplasm without fusing into lysosomes [38]. Therefore, PBAE/DNA polyplexes internalized via caveolae and macropinocytosis would not experience endosomal entrapment and lysosomal degradation, which greatly contributed to their relatively high gene transfection efficiencies. In support of such statement, we noticed that the internalized polyplexes containing YOYO-1-DNA were largely separated from Lysotracker Red-stained endosomes/lysosomes after 4-h uptake process, which substantiated that they effectively avoided endosomal/lysosomal entrapment (Fig. 6C).
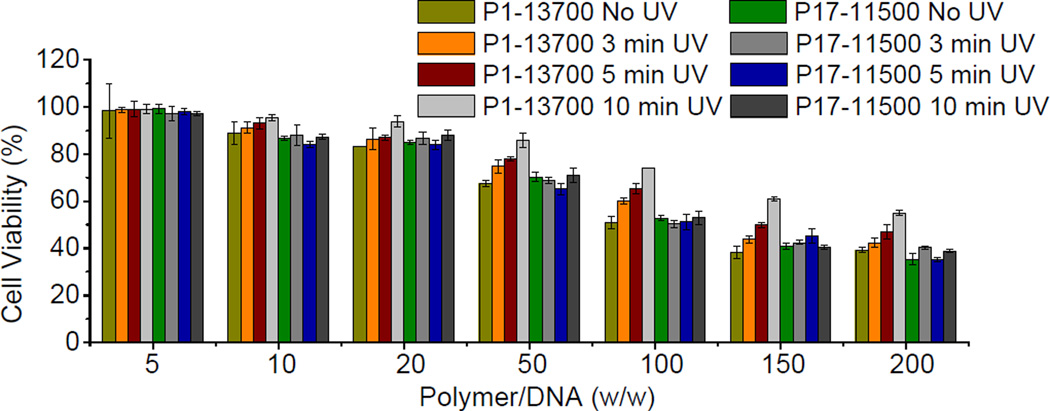
3.7. Cytotoxicity
Cationic polymers with lower MWs are much easier to be expelled from the biological membranes because of fewer contact points with the cell components for each individual polymeric chain, and accordingly exhibit lower cytotoxicity than those with higher MWs [17, 41]. Motivated by such statement, we next evaluated whether light-triggered backbone degradation would alleviate the cytotoxicity of PBAEs. In accordance with the transfection process, cells were treated with polyplexes at various polymer/DNA weight ratios for 4 h, irradiated with UV light (λ = 365 nm, 20 mW/cm2) for different time, further cultured for 48 h, and analyzed for cell viability using the MTT assay. As shown in Fig. 7, UV irradiation at the post-transfection state significantly reduced the cytotoxicity of PBAEs, and longer irradiation time yielded further improved cell viability, which was attributed to promoted polymer degradation into smaller fragments. Such observation thus validated our proposed design strategy to improve the cell tolerability of PBAEs by post-transfection light exposure.
Fig. 7.
Viabilities of HeLa cells transfected with polyplexes at various polymer/DNA weight ratios and irradiated with UV light (λ = 365 nm, 20 mW/cm2) for different time periods (n = 3). Cells were treated with polyplexes for 4 h, irradiated with UV light, and further cultured for 48 h before viability assessment using the MTT assay.
4. Conclusion
In summary, we developed a strategy to promote intracellular DNA release as well as reduce polycation toxicity by controlling polymer degradation using external triggers. With this strategy, a library of PBAEs with built-in trigger-responsive domains in the backbone were developed which undergo instantaneous degradation upon external stimuli to trigger DNA unpackaging and self-diminish the cytotoxicity at the post-transfection state. By provoking these multiple intracellular responses, transfection efficiencies of PBAEs were greatly improved to outperform commercial reagent LPF. Such strategy provides an effective tool in overcoming the multiple cellular barriers against polycation-mediated gene transfer, and will thus provide insights into the design of non-viral gene delivery vectors. Although UV light was used as the external trigger in this proof-of-concept study, other responsive domains will be designed in future studies to allow response to non-toxic stimuli, such as NIR light.
Supplementary Material
Acknowledgement
J.C. acknowledges support from the NSF (CHE-1153122) and the NIH (NIH Director's New Innovator Award 1DP2OD007246 and 1R21EB013379).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Candolfi M, Xiong W, Yagiz K, Liu C, Muhammad AK, Puntel M, et al. Gene therapy-mediated delivery of targeted cytotoxins for glioma therapeutics. Proc Natl Acad Sci U S A. 2010;107:20021–20026. doi: 10.1073/pnas.1008261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang JL, Tang GP, Shen J, Hu QL, Xu FJ, Wang QQ, et al. A gene nanocomplex conjugated with monoclonal antibodies for targeted therapy of hepatocellular carcinoma. Biomaterials. 2012;33:4597–4607. doi: 10.1016/j.biomaterials.2012.02.045. [DOI] [PubMed] [Google Scholar]
- 3.Cho SK, Pedram A, Levin ER, Kwon YJ. Acid-degradable core–shell nanoparticles for reversed tamoxifen-resistance in breast cancer by silencing manganese superoxide dismutase (MnSOD) Biomaterials. 2013;34:10228–10237. doi: 10.1016/j.biomaterials.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin L, Song Z, Qu Q, Kim KH, Zheng N, Yao C, et al. Supramolecular self-assembled nanoparticles mediate oral delivery of therapeutic TNF-α siRNA against systemic inflammation. Angew Chem Int Ed. 2013;52:5757–5761. doi: 10.1002/anie.201209991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CK, Law WC, Aalinkeel R, Nair B, Kopwitthaya A, Mahajan SD, et al. Well-defined degradable cationic polylactide as nanocarrier for the delivery of siRNA to silence angiogenesis in prostate cancer. Adv Healthc Mater. 2012;1:751–761. doi: 10.1002/adhm.201200094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia FJ, Wilson KD, Sun N, Gupta DM, Huang M, Li ZJ, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7 doi: 10.1038/nmeth.1426. 197-U46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li ZX, Dullmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 10.Baum C, Kustikova O, Modlich U, Li ZX, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 11.Chu DSH, Schellinger JG, Bocek MJ, Johnson RN, Pun SH. Optimization of Tet1 ligand density in HPMA-co-oligolysine copolymers for targeted neuronal gene delivery. Biomaterials. 2013;34:9632–9637. doi: 10.1016/j.biomaterials.2013.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei H, Volpatti LR, Sellers DL, Maris DO, Andrews IW, Hemphill AS, et al. Dual responsive, stabilized nanoparticles for efficient in vivo plasmid delivery. Angew Chem Int Edit. 2013;52:5377–5381. doi: 10.1002/anie.201301896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C-K, Jones CH, Mistriotis P, Yu Y, Ma X, Ravikrishnan A, et al. Poly(ethylene glycol)-block-cationic polylactide nanocomplexes of differing charge density for gene delivery. Biomaterials. 2013;34:9688–9699. doi: 10.1016/j.biomaterials.2013.08.063. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X, Yin LC, Ding JY, Tang C, Gu SH, Yin CH, et al. Thiolated trimethyl chitosan nanocomplexes as gene carriers with high in vitro and in vivo transfection efficiency. J Control Release. 2010;144:46–54. doi: 10.1016/j.jconrel.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Teo PY, Yang C, Hedrick JL, Engler AC, Coady DJ, Ghaem-Maghami S, et al. Hydrophobic modification of low molecular weight polyethylenimine for improved gene transfection. Biomaterials. 2013;34:7971–7979. doi: 10.1016/j.biomaterials.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Song YB, Wang HY, Zeng XA, Sun YX, Zhang XZ, Zhou JP, et al. Effect of molecular weight and degree of substitution of quaternized cellulose on the efficiency of gene transfection. Bioconjugate Chem. 2010;21:1271–1279. doi: 10.1021/bc100068f. [DOI] [PubMed] [Google Scholar]
- 17.Yin LC, Ding JY, He CB, Cui LM, Tang C, Yin CH. Drug permeability and mucoadhesion properties of thiolated trimethyl chitosan nanoparticles in oral insulin delivery. Biomaterials. 2009;30:5691–5700. doi: 10.1016/j.biomaterials.2009.06.055. [DOI] [PubMed] [Google Scholar]
- 18.Shim MS, Kwon YJ. Controlled cytoplasmic and nuclear localization of plasmid DNA and siRNA by differentially tailored polyethylenimine. J Control Release. 2009;133:206–213. doi: 10.1016/j.jconrel.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Mol Ther. 2005;11:426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Edit. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 21.Lynn DM, Anderson DG, Putnam D, Langer R. Accelerated discovery of synthetic transfection vectors: Parallel synthesis and screening of degradable polymer library. J Am Chem Soc. 2001;123:8155–8156. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- 22.Lynn DM, Langer R. Degradable poly(beta-amino esters): Synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 23.Akinc A, Lynn DM, Anderson DG, Langer R. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. J Am Chem Soc. 2003;125:5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 24.Anderson DG, Peng WD, Akinc A, Hossain N, Kohn A, Padera R, et al. A polymer library approach to suicide gene therapy for cancer. P Natl Acad Sci USA. 2004;101:16028–16033. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeney M, Ong SG, Padilla A, Yao ZY, Goodman S, Wu JC, et al. Development of poly(beta-amino ester)-based biodegradable nanoparticles for nonviral delivery of minicircle DNA. Acs Nano. 2013;7:7241–7250. doi: 10.1021/nn402657d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunshine JC, Sunshine SB, Bhutto I, Handa JT, Green JJ. Poly(beta-amino ester)-nanoparticle mediated transfection of retinal pigment epithelial cells in vitro and in vivo. Plos One. 2012;7:e37543. doi: 10.1371/journal.pone.0037543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang YH, Zugates GT, Peng WD, Holtz D, Dunton C, Green JJ, et al. Nanoparticle-delivered suicide gene therapy effectively reduces ovarian tumor burden in mice. Cancer Res. 2009;69:6184–6191. doi: 10.1158/0008-5472.CAN-09-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozielski KL, Tzeng SY, Green JJ. A bioreducible linear poly(beta-amino ester) for siRNA delivery. Chem Commun. 2013;49:5319–5321. doi: 10.1039/c3cc40718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunshine JC, Peng DY, Green JJ. Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol Pharmaceut. 2012;9:3375–3383. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin LC, Tang HY, Kim KH, Zheng N, Song ZY, Gabrielson NP, et al. Light-responsive helical polypeptides capable of reducing toxicity and unpacking DNA: toward nonviral gene delivery. Angew Chem Int Edit. 2013;52:9182–9186. doi: 10.1002/anie.201302820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin L, Song Z, Kim KH, Zheng N, Gabrielson NP, Cheng J. Non-viral gene delivery via membrane-penetrating, mannose-targeting supramolecular self-assembled nanocomplexes. Adv Mater. 2013;25:3063–3070. doi: 10.1002/adma.201205088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bishop CJ, Ketola TM, Tzeng SY, Sunshine JC, Urtti A, Lemmetyinen H, et al. The effect and role of carbon atoms in poly(beta-amino ester)s for DNA binding and gene delivery. J Am Chem Soc. 2013;135:6951–6957. doi: 10.1021/ja4002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eltoukhy AA, Siegwart DJ, Alabi CA, Rajan JS, Langer R, Anderson DG. Effect of molecular weight of amine end-modified poly(beta-amino ester)s on gene delivery efficiency and toxicity. Biomaterials. 2012;33:3594–3603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akinc A, Anderson DG, Lynn DM, Langer R. Synthesis of poly(beta-amino ester)s optimized for highly effective gene delivery. Bioconjugate Chem. 2003;14:979–988. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- 35.Midoux P, Pichon C, Yaouanc JJ, Jaffres PA. Chemical vectors for gene delivery: a current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Brit J Pharmacol. 2009;157:166–178. doi: 10.1111/j.1476-5381.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang SN, Su J, Tong HJ, Yang F, Tong WX, Yuan WE, et al. Biscarbamate cross-linked low molecular weight PEI for delivering IL-1 receptor antagonist gene to synoviocytes for arthritis therapy. Biomaterials. 2012;33:6520–6532. doi: 10.1016/j.biomaterials.2012.05.044. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed M, Narain R. The effect of molecular weight, compositions and lectin type on the properties of hyperbranched glycopolymers as non-viral gene delivery systems. Biomaterials. 2012;33:3990–4001. doi: 10.1016/j.biomaterials.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, et al. The effect of particle design on cellular internalization pathways. P Natl Acad Sci USA. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang H, Yin L, Kim KH, Cheng J. Helical poly(arginine) mimics with superior cell-penetrating and molecular transporting properties. Chem Sci. 2013;4:3839–3844. doi: 10.1039/C3SC51328A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin L, Song Z, Kim KH, Zheng N, Tang H, Lu H, et al. Reconfiguring the architectures of cationic helical polypeptides to control non-viral gene delivery. Biomaterials. 2013;34:2340–2349. doi: 10.1016/j.biomaterials.2012.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao Z, Ma L, Jiang Y, Yan M, Gao C, Shen J. N,N,N-trimethylchitosan chloride as a gene vector: synthesis and application. Macromol Biosci. 2007;7:855–863. doi: 10.1002/mabi.200700015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.