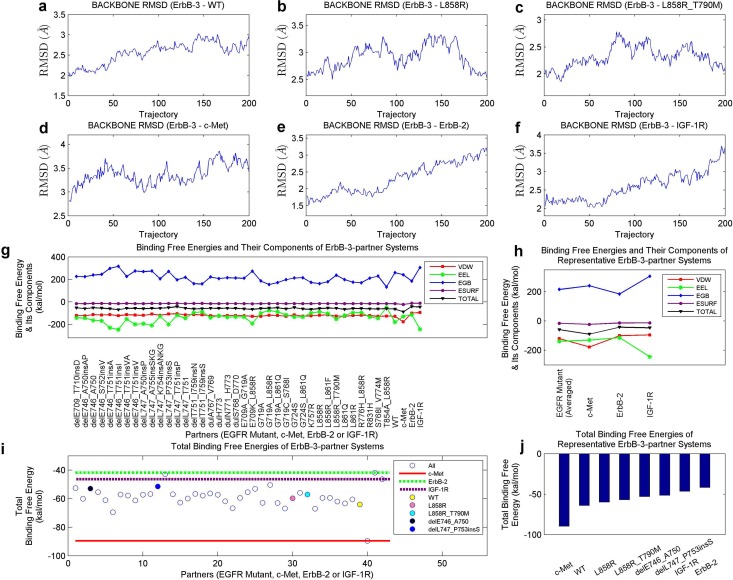
Fig 10. Computational results for ErbB-3-partner systems.
(a) to (f), Respective backbone RMSD curves of ErbB-3—WT, ErbB-3—L858R, ErbB-3—L858R_T790M, ErbB-3—c-Met, ErbB-3—ErbB-2 and ErbB-3—IGF-1R systems, in the 2-ns production MD simulations. (g) Binding free energies and their components of ErbB-3-partner systems. (h) Binding free energies and their components of ErbB-3—EGFR mutant (averaged), ErbB-3—c-Met, ErbB-3—ErbB-2 and ErbB-3—IGF-1R systems. (i) Total binding free energy comparison among ErbB-3-partner systems. Binding free energies of ErbB-3—c-Met, ErbB-3—ErbB-2 and ErbB-3—IGF-1R systems are labeled with lines, and those of systems involving WT EGFR, L858R, L858R_T790, delE746_A750 and delL747_P753insS are marked with solid spheres. (j) Ranked total binding free energies of representative ErbB-3-partner systems.