Fig 2. Characterization of the interaction of PARP-1 with the -20/-10 EPHX1 promoter region.
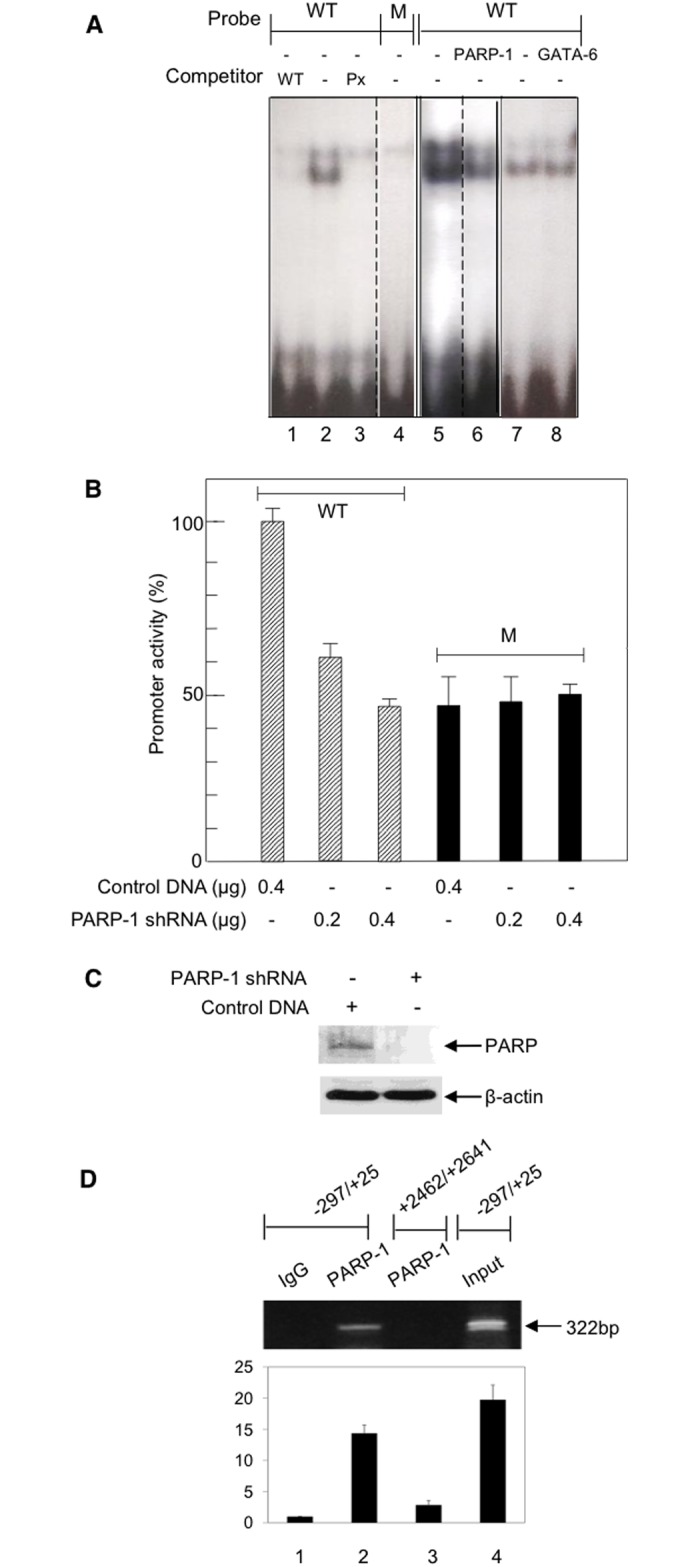
A. EMSA analysis was performed with a HepG2 nuclear protein extract and 32P-labeled oligonucleotides (-28/-4) containing the WT PARP-1 sequence in the absence and presence of a 50-fold excess of unlabeled oligonucleotide WT competitor (lane 2,1), and in the presence of a 50-fold excess of an oligonucleotide (Px) of unrelated sequence that can also binds PARP-1 (lane 3). EMSA was performed with the labeled -17 mutated oligonucleotide (lane 4). Nuclear extracts were preincubated with a PARP-1 antibody (lane 5,6) or a GATA-6 antibody (lane 7,8) prior to EMSA analysis. B. The effect of PARP-1 shRNA on the promoter activity of the EPHX1 -8-/+25 construct. Values are the mean +/- S.D. of independent experiments (n = 3–5) performed in triplicate. C. Western blot of PARP-1 levels following treatment with 0.4 μg PARP-1 shRNA. D. The interaction of PARP-1 with the EPHX1 proximal promoter as assessed by ChIP analysis. Crosslinked protein-DNA complexes were incubated with anti-PARP-1 antibody, isolated by protein A-Sepharose beads and DNA corresponding to the EPHX1–297/+25 and EPHX1 +2462/+2641 which does not contain the PARP-1 binding site were analyzed by PCR. The band observed for PARP-1 in the EPHX1 promoter region (lane 2). Immunoprecipatation with IgG (lane 1). PCR analysis with primers to the +2462/+2641 region (lane 3). Gels were scanned and quantitated using UN-SCAN-IT gel automated digitizing system. Values are the result of 3 independent experiments.
