Figure 4. Exocytosis of BDNF vesicles is controlled by two different SNAPs.
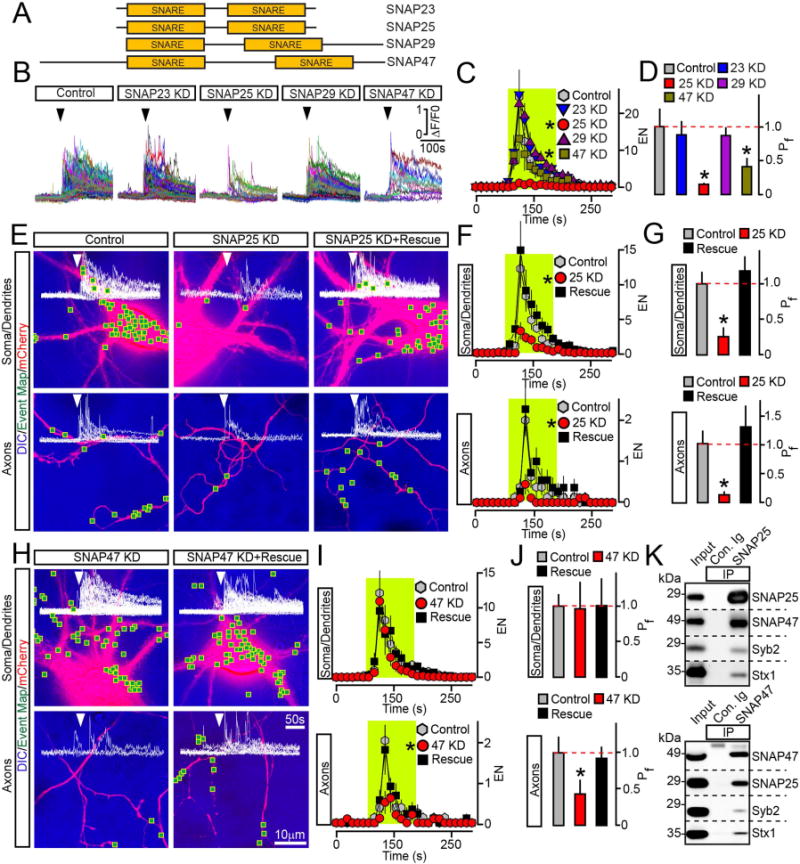
(A) Domain structures of vertebrate SNAREs of the SNAP family.
(B to D) Outcomes of global ablation of SNAPs on BDNF release. Cortical cultures were co-infected at DIV5 with viruses expressing BDNF-pHluorin and two shRNAs that target non-overlapping sequences of specific SNAP isoforms (KD). Exocytosis was monitored at DIV15 by TIRF imaging. (B) Sample traces reflecting all fusion evens in control and SNAP-deficient neurons. Arrows mark times of depolarization with KCl. (C) Averaged EN, plotted as a function of time. (D) Mean Pf across neuronal somas and processes. Control, n = 10; SNAP23 KD, n = 8; SNAP25 KD, n = 8; SNAP29 KD, n = 7; SNAP47 KD, n = 8. * p < 0.01 and < 0.05 for SNAP25 and SNAP47, respectively (ANOVA).
(E to J) Cell-autonomous effects of SNAP ablation on BDNF release. Exocytosis was monitored by TIRF microscopy in isolated neurons in DIV15 cultures that were sparsely co-infected with viruses encoding BDNF-pHluorin, mCherry and shRNAs against indicated SNAPs without or with corresponding rescue cDNAs. Panels show aligned DIC and mCherry images with mapped sites of depolarization-induced fusion, actual events of exocytosis in each site, EN plotted as a function of time, and Pf. n = 9–11/group. (E to G) Examples of exocytosis, averaged EN, and Pf in different subcellular compartments of control and SNAP25-deficient neurons. * p < 0.01 (ANOVA). (H to J) Examples of exocytosis, mean EN, and Pf in SNAP47-deficient neurons. * p < 0.05 (ANOVA). Control groups apply to both SNAP isoforms. Quantifications are represented as Mean ± S.E.M. See also Supplemental Figures 5 and 6.
(K) Solubilized proteins were co-immunoprecipitated (IP) from mouse forebrain extracts with control antibody (Ig) or antibodies against SNAP25 (top) or SNAP47 (bottom). Attached complexes were examined by immunoblotting with antibodies to SNAP25, SNAP47, Syb2 and Syntaxin1. Equal amounts of total protein were applied to each lane.
