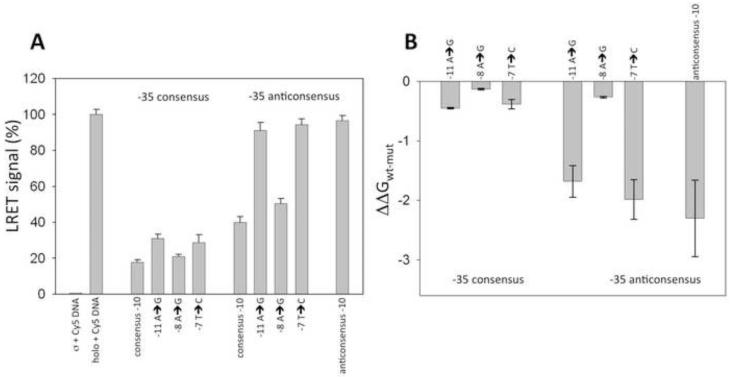
Figure 2.
LRET-based determination of relative binding affinity of fork junction DNA constructs. (A) Sensitized acceptor signal (LRET) in the absence of unlabeled competitor DNA (first two bars) and in the presence of competitor fork junction DNA’s containing indicated mutations. Data were normalized to the signal observed for the holoenzyme in the presence of Cy5 labeled R1 DNA construct (second bar). (B) Relative binding affinity of the fork junction DNA constructs calculated from the data shown in panel A using eq. 1 and 2.