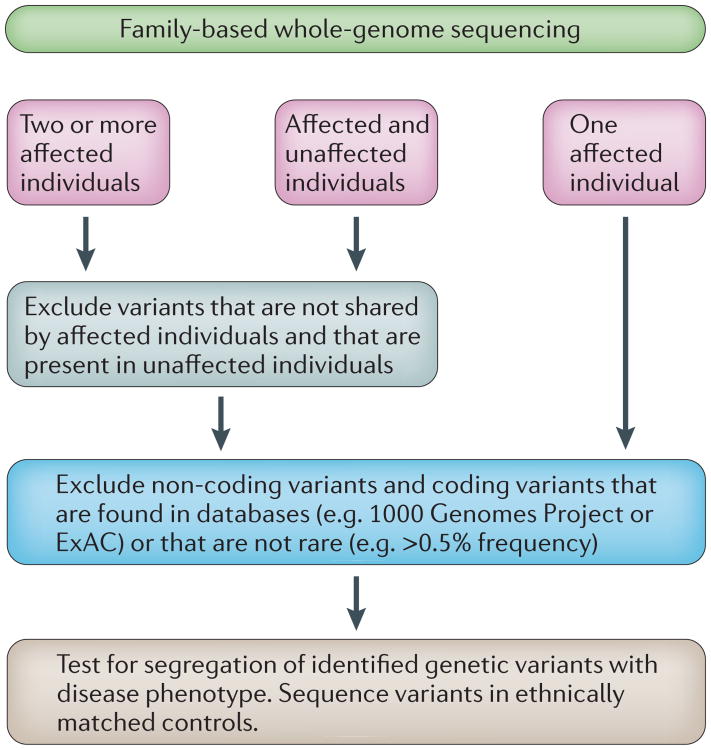
Figure 1. Workflow for the whole-genome sequencing filtering approach in human family data.
Usually, one, two or more affected individuals, or affected and unaffected individuals, in a family have their genomes or exomes sequenced. Variants that are not predicted to be nonsense, missense or splice-site variants are usually excluded from further analyses because it is unlikely that they are causal. When the mode of inheritance of a disease is known, this information can be used to aid the selection of variants. For example, for an autosomal dominant disease, the affected pedigree member's sequence data should display a heterozygous causal variant. Sequence data on additional pedigree members can help to reduce the number of variants that could potentially be disease causing. A final filtering step is performed in which those variants that are present in the databases dbSNP, 1000 Genomes, ExAC and Exome Variant Server are excluded. Additionally, bioinformatic tools, such as Polyphen-2 (Ref. 102), and measures of conservation, for example, PhyloP103, are often used to predict whether a variant is deleterious and therefore likely to be disease causing. Even after filtering steps, there may be many variants that need to be followed up in the remaining family members to elucidate whether the variant (or variants) segregate with the disease phenotype. If the family is from a population that is not represented in databases, then ethnically matched controls need to be sequenced to evaluate the frequency of the variant (or variants).