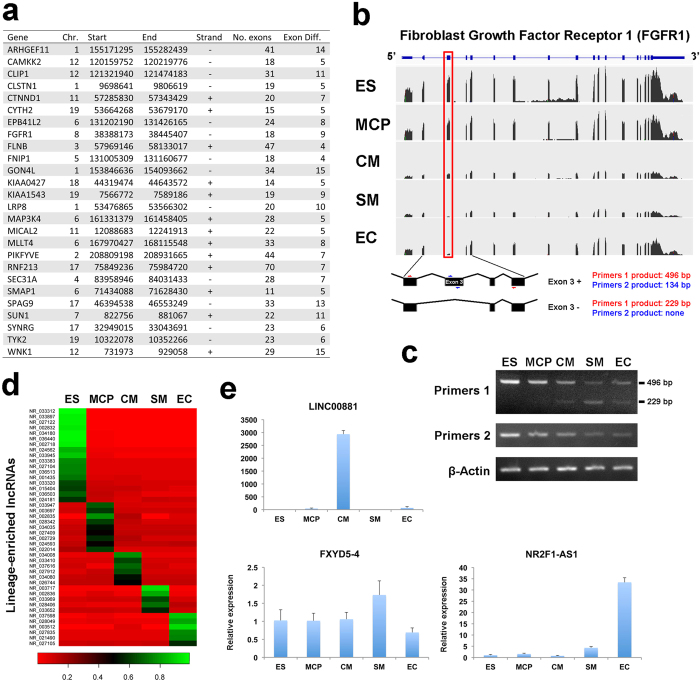
Figure 7.
GEPA predicted differentiation-associated alternative RNA splicing and lineage-enriched long non-coding RNAs during human cardiovascular differentiation. (a) Genes with exon changes detected by RNA-seq during human cardiovascular differentiation. Locations of the genes in Hg18 human genome assembly were shown along with total exon number (No. exons) and number of exons with changes (Exon diff.). (b) Differential use of the exon 3 of FGFR1 during human cardiovascular differentiation. Stacks of sequencing reads in the five lineages were shown. Distances of the exons are NOT the actual intron lengths. Change in the expression of exon 3 was highlighted by a red frame. Primers to verify the expression of the exon 3 were indicated by red or blue arrows. (c) Semi-quantitative RT-PCR validation of the exon 3 usage during human cardiovascular differentiation. (d) Lineage-enriched long non-coding RNAs during human cardiovascular differentiation identified by GEPA. To indicate the lineage-specificity, the relative expression was shown as percent in the sum of all the cell types in the heatmap. (e) qRT-PCR validation of the lineage-enriched expressions of lncRNAs.