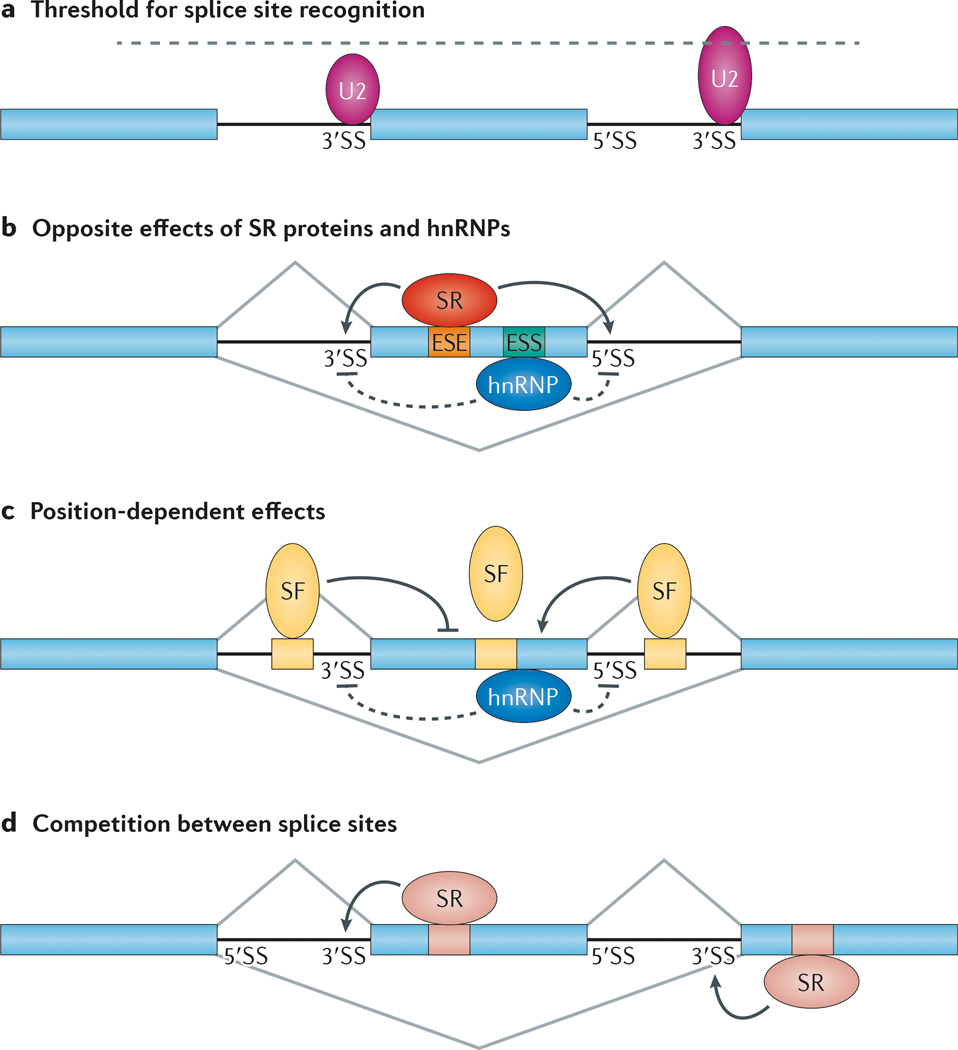
Figure 3. Rules for context-dependent and position-sensitive regulation of alternative splicing.
a | In general, the alternative exon (in the middle) is associated with weak splice site signals, which render partial selection by core components of the splicing machinery. A small U2 small nuclear ribonucleoprotein (snRNP) is illustrated to show its inefficient interaction with the alternative splice site. The flanking sites, which are strongly recognized by the splicing machinery, are engaging in competition with the alternative splice sites. A large U2 snRNP is illustrated to show its strong interaction with the flanking 3′ splice site (3′SS). According to the threshold rule, a partially compromised splicing machinery would selectively affect the alternative splice sites relative to the competing sites, thus leading to induced exon skipping. b | Exon-bound SR proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) have opposite effects on the recognition of the adjacent splice sites by the core splicing machinery. c | For many sequence-specific splicing factors (SFs) — for example, NOVA — their binding on exons generally inhibits the selection of the alternative exon. They are inhibitory when they are bound to an upstream intronic region but enhance the selection of the alternative exon when bound to a downstream intronic region. The mechanism for the polarity effect remains to be fully understood. d | Enhanced recognition of the alternative splice site induces exon inclusion, whereas increased recognition of the flanking competing splice site causes exon skipping. ESE, exonic splicing enhancer; ESS, exonic splicing silencer.