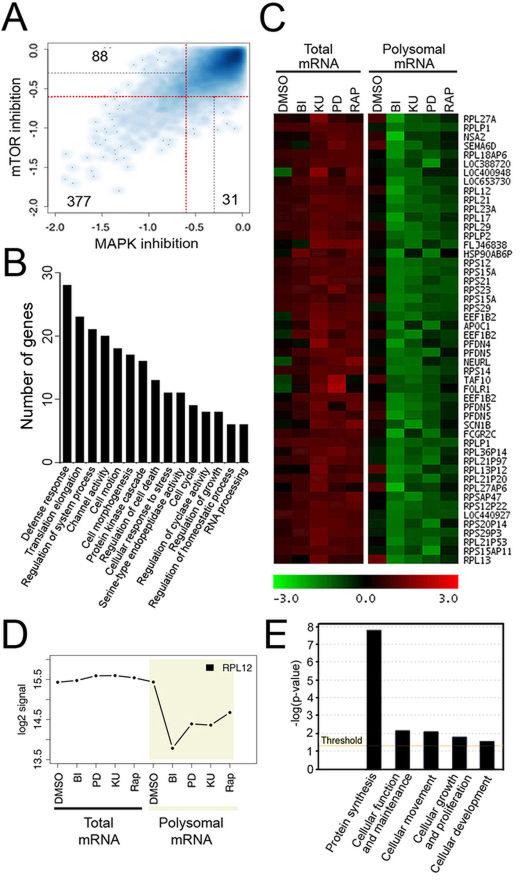
Fig. 5.
ERK/RSK signalling promotes translation of growth-related mRNAs in melanoma. (A) Scatter plot of mRNAs log2-coefficients in polysomal mRNA from serum-starved Colo829 cells treated with mTOR (rapamycin or KU-0063794) or MAPK (BI-D1870 or PD184352) pathway inhibitors. Red and grey lines represent cutoffs of 1.5-fold (log2-coefficient=−0.58) and 1.25-fold (log2-coefficient=−0.32) decrease, respectively. mRNAs regulated by both types of inhibitors are found in the bottom-left quadrant delimited by two red lines whereas mRNAs regulated by only one inhibitor class are found at the top-left (MAPK) or bottom-right of the plot (mTOR). (B) Gene categories found as enriched by DAVID analysis in the list of downregulated mRNAs identified as translationally repressed by both mTOR and MAPK inhibitors (treatment effect<1.5-fold decrease and pvalue <0.05). (C) K-Means clustering of downregulated mRNAs in the presence of both mTOR and MAPK inhibitors led to the identification of a cluster whose mRNAs are translationally repressed. mRNAs and sample class are presented vertically and horizontally, respectively. Green and red represent respectively a relative decrease and a relative increase in mRNA abundance compared to the mRNA mean abundance. (D) Profile of a selected mRNA identified by clustering with log2 signal on the y-axis and the different sample classes on the x-axis. (E) Enrichment analysis using Ingenuity Pathways Analysis of mRNAs from identified clusters.