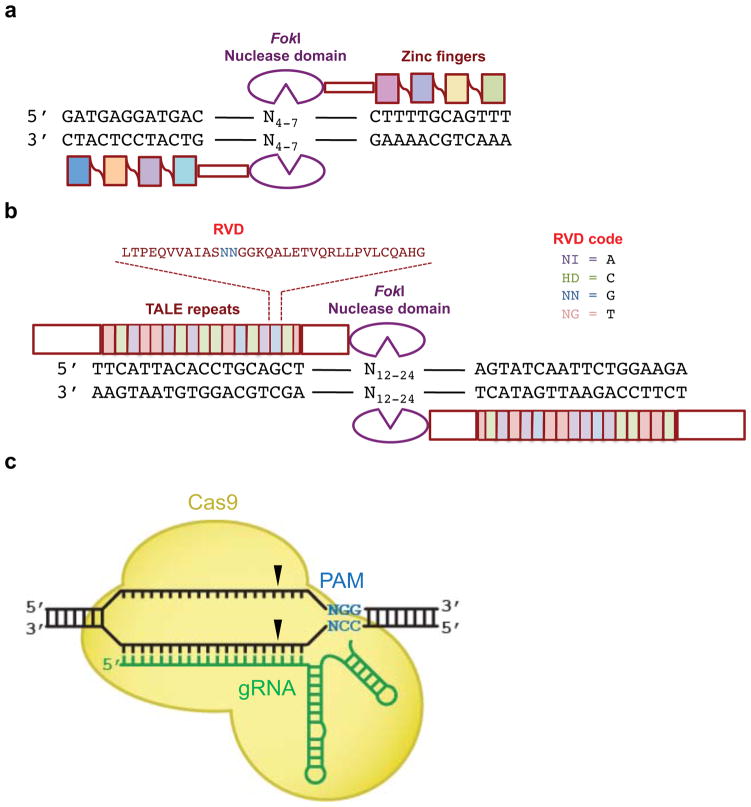
Figure 3. Architecture of ZFN, TALEN, and Cas9 programmable nucleases.
(a) A ZFN monomer is a fusion of a FokI nuclease cleavage domain (purple) to (typically) four adjoined zinc-fingers each targeting three base pairs for a total of 12 base pairs recognized. Two different ZFNs bind their corresponding half-sites, allowing FokI dimerization and DNA cleavage between the half-sites. (b) A TALEN monomer contains an N-terminal domain followed by an array of TALE repeats (filled boxes), a C-terminal domain, and a FokI nuclease cleavage domain (purple). The 12th and 13th amino acids (the RVD, red) of each TALE repeat recognize a specific DNA base pair. Two different TALENs bind their corresponding half-sites, allowing FokI dimerization and DNA cleavage between the half-sites. (c) Cas9 protein (yellow) binds to target DNA in complex with a single guide RNA (sgRNA, green). The S. pyogenes Cas9 protein and sgRNA complex recognizes the PAM sequence NGG (blue). Black triangles indicate the cleavage points in the target DNA three bases from the PAM on both DNA strands.