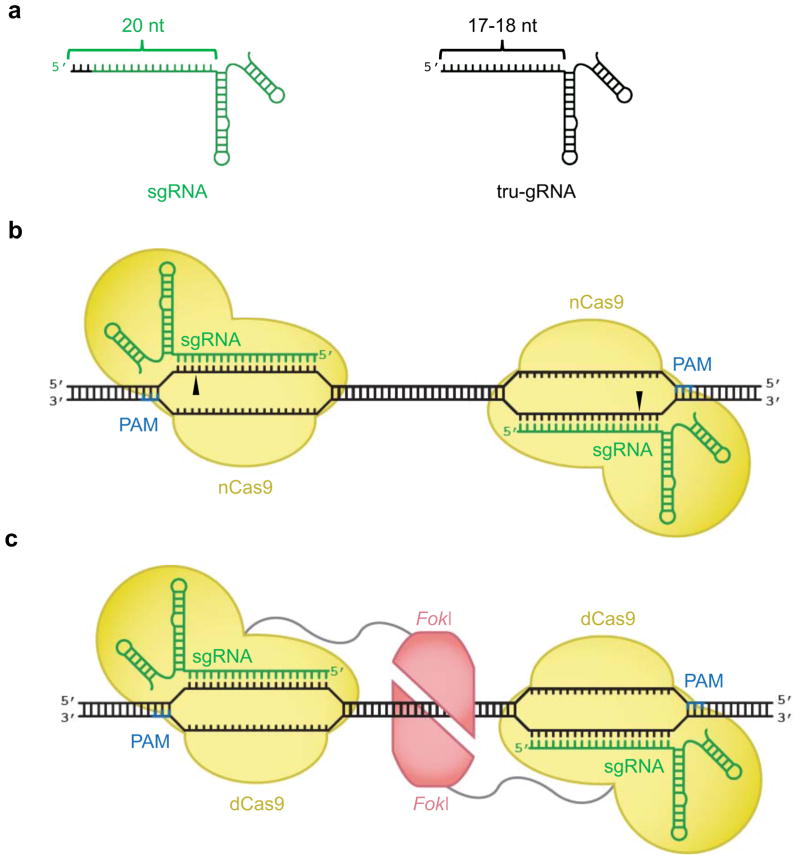
Figure 4. Engineered Cas9 components with improved DNA cleavage specificity.
(a) A truncated guide RNA (tru-gRNA, right), contains 17–18 base pairs of complementarity to its DNA target site, rather than 20 base pairs in a canonical sgRNA (left). The base pairs in the sgRNA that are not present in the tru-gRNA are colored black. (b) Mutant Cas9 proteins that cleave only a single strand of dsDNA (nCas9) can be targeted to opposite strands of adjacent sites as pairs to cause double strand breaks. (c) Monomers of FokI nuclease (red) fused to catalytically inactive dCas9 bind to separate sites within a target locus. Only adjacently bound FokI-dCas9 monomers can assemble a catalytically active FokI nuclease dimer, triggering dsDNA cleavage.