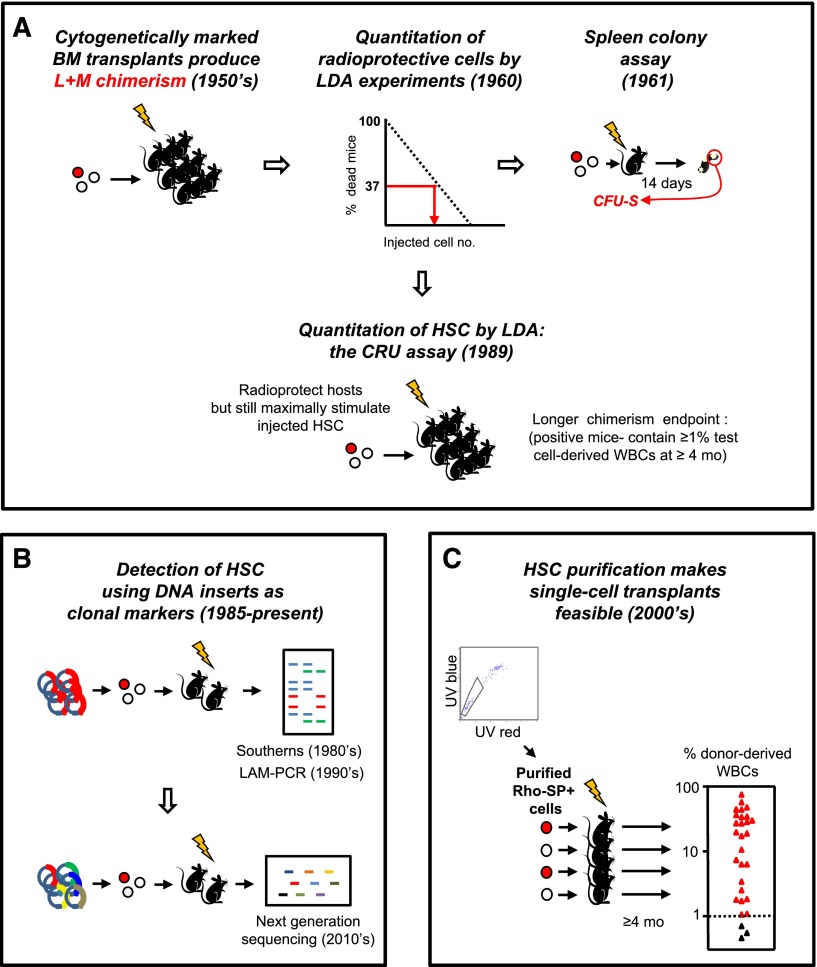
Figure 1.
Historical sequence of methods used to detect and quantify mouse HSCs in vivo. (A) Development of LDA approaches to identify the transplantable cells that can rescue mice permanently from radiation-induced lethality by regenerating the inactivated blood-forming system of the host. (B) Genetic approaches to track HSCs by detection of their long-lived clonal outputs in transplanted recipients. Random sites of vector integration into the DNA of the regenerated progeny of transduced transplanted cells were the first unique DNA identifiers used.3,4 More recently, uptake of a single vector encoding a short unique “barcode” sequence from a diverse vector library has been used as a clonal tracking strategy.94-97 (C) Advances in LTRC purification enabling single-cell transplants to reveal the diversity of long-term clonal white blood cell outputs of individual HSCs as previously suggested by limiting dilution transplants and vector-marking experiments. Data shown are for purified Rho−SP+ LTRCs (HSCs) adapted from Uchida et al5 (with permission from Experimental Hematology.) BM, bone marrow; L+M, lymphoid + myeloid; mo, months; LAM-PCR, linear amplification mediated polymerase chain reaction; SP, side population; UV, ultraviolet; WBCs, white blood cells.