Figure 1.
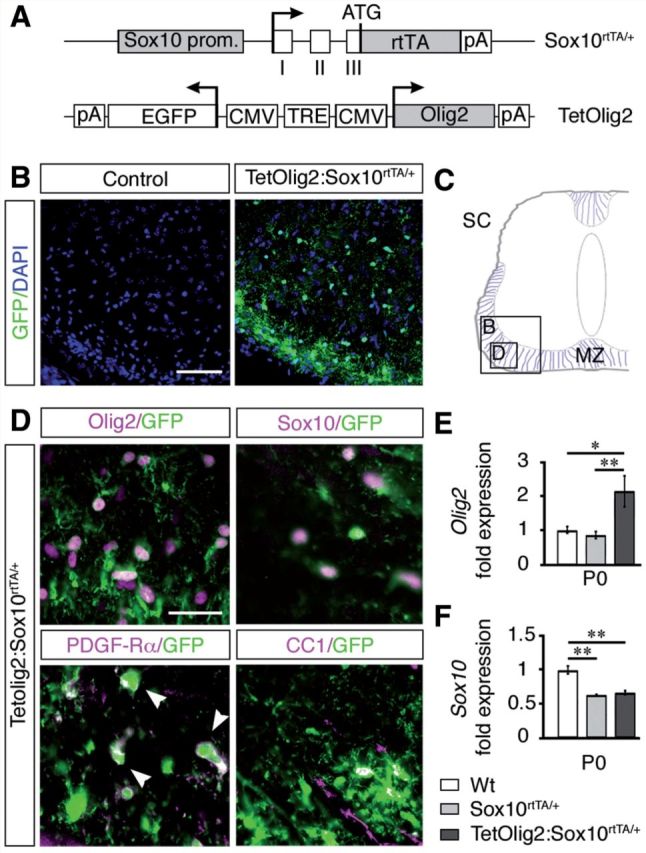
Generation of TetOlig2:Sox10rtTA/+ mouse line. (A) Schematic representation of Sox10rtTA/+ and TetOlig2 alleles that together induce Olig2 overexpression in Sox10+ cells upon doxycycline treatment. Sox10 prom = Sox10 promoter region; open boxes = exons I–III of the Sox10 gene; rtTA = coding sequences for the reverse tetracycline-controlled transactivator; pA = polyadenylation sites; TRE = tetracycline responsive element; CMV = minimal promoter of cytomegalovirus immediate early genes; Olig2 = coding sequence of the Olig2 gene; EGFP = coding sequence of the enhanced green fluorescent protein; ATG = start codon of the Sox10 gene. Arrows indicate transcription starts. (B) GFP immunohistochemistry (green) in control (wild-type or Sox10rtTA/+) and TetOlig2:Sox10rtTA/+ spinal cord at P0. Sections are counterstained with DAPI (blue). Note that GFP was only detected in TetOlig2:Sox10rtTA/+ mice. (C) Schematic representation of the spinal cord (SC) illustrating the marginal zone (MZ) and regions shown in B and D (open boxes). (D) Immunolabellings for GFP (green), Olig2, Sox10, PDGFRA and CC1 (all in red) on spinal cord sections of TetOlig2:Sox10rtTA/+. GFP was detected in Olig2+, Sox10+, PDGFRA+ and CC1+ oligodendroglial cells. (E and F) Quantitative reverse transcriptase PCR for Olig2 and Sox10 (at least n = 3 for each genotype). (E) In P0 TetOlig2:Sox10rtTA/+ mice (dark grey bars), Olig2 expression was significantly increased with respect to wild-type (Wt) (white bars, P = 0.012) and Sox10rtTA/+ littermates (light grey bars, P = 0.009). (F) Sox10 expression level was reduced by half in Sox10rtTA/+ and TetOlig2:Sox10rtTA/+ with respect to wild-type (P = 0.002 and P = 0.0017, respectively). Scale bars: B = 75 µm; D = 20 µm.
