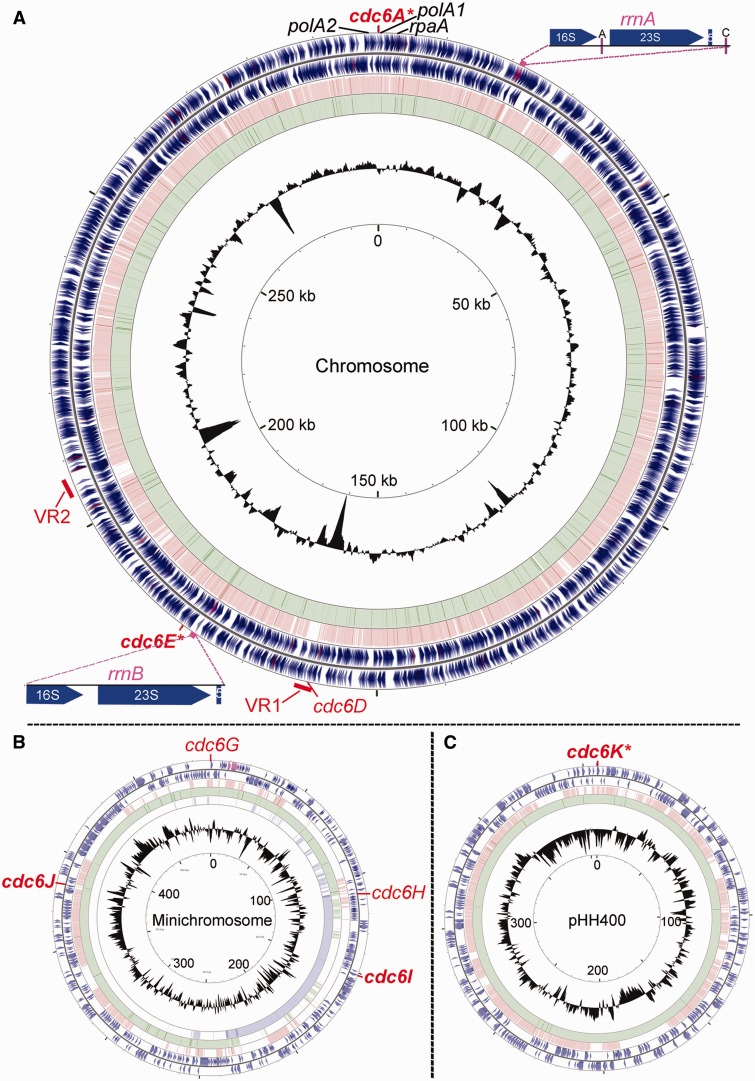
Fig. 1.—
Comparative genomic analysis of the replication origins in Haloarcula species. (A) The chromosome. From the outside inward: The first and second circles indicate the annotated protein-coding regions on the plus and minus strands in H. hispanica ATCC 33960. The third and fourth circles represent the BLASTN hits of the chromosome of H. hispanica ATCC 33960 with those of two other Haloarcula genomes: H. marismortui ATCC 43049 and H. hispanica N61, respectively. The innermost circle shows a plot of the GC content along the chromosome of H. hispanica ATCC 33960. The two large variable regions (VR1-2, red) and the positions of two rrn operons are indicated. The two rrn operons are both directed away from the origins. (B and C) Minichromosome and megaplasmid pHH400. The circles in B and C are the same as in A with the exception that the H. hispanica N61 minichromosome was divided into two replicons: The minichromosome and pHH126 (the fourth and fifth circles in B). In all three replicons, the cdc6 genes associated with the putative replication origins in H. hispanica are indicated, and those with active origins are indicated in bold. The conserved ori-cdc6 origins in Haloarcula species, namely oriC1-cdc6A and oriC2-cdc6E on the chromosome and oriP-cdc6K on PHH400, are highlighted with star signs. GC, Guanine-Cytosine.